Abstract
Inflammatory bowel diseases present with elevated levels of intestinal epithelial cell (IEC) death, which compromises the gut barrier, activating immune cells and triggering more IEC death. The endogenous signals that prevent IEC death and break this vicious cycle, allowing resolution of intestinal inflammation, remain largely unknown. Here we show that prostaglandin E2 signalling via the E-type prostanoid receptor 4 (EP4) on IECs represses epithelial necroptosis and induces resolution of colitis. We found that EP4 expression correlates with an improved IBD outcome and that EP4 activation induces a transcriptional signature consistent with resolution of intestinal inflammation. We further show that dysregulated necroptosis prevents resolution, and EP4 agonism suppresses necroptosis in human and mouse IECs. Mechanistically, EP4 signalling on IECs converges on receptor-interacting protein kinase 1 to suppress tumour necrosis factor-induced activation and membrane translocation of the necroptosis effector mixed-lineage kinase domain-like pseudokinase. In summary, our study indicates that EP4 promotes the resolution of colitis by suppressing IEC necroptosis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA-seq data that support the findings of this study have been deposited in the Array Express service of the Molecular Biology Laboratory–European Bioinformatics Institute under accessions E-MTAB-9850 and E-MTAB-9820. Previously published sequencing data that were re-analysed here are available under accession codes GSE1122360, GSE5794561, GSE10914262, GSE12868263, GSE1687964, GSE5907165, GSE11693666, GDS518214 and E-MTAB-647667. All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Yukawa, M. et al. Systemic and local evidence of increased Fas-mediated apoptosis in ulcerative colitis. Int J. Colorectal Dis. 17, 70–76 (2002).
Iwamoto, M., Koji, T., Makiyama, K., Kobayashi, N. & Nakane, P. K. Apoptosis of crypt epithelial cells in ulcerative colitis. J. Pathol. 180, 152–159 (1996).
Jensen, S., Seidelin, J. B., LaCasse, E. C. & Nielsen, O. H. SMAC mimetics and RIPK inhibitors as therapeutics for chronic inflammatory diseases. Sci. Signal. 13, eaax8295 (2020).
Schett, G. & Neurath, M. F. Resolution of chronic inflammatory disease: universal and tissue-specific concepts. Nat. Commun. 9, 3261 (2018).
Rogler, G. Resolution of inflammation in inflammatory bowel disease. Lancet Gastroenterol. Hepatol. 2, 521–530 (2017).
Ting, A. T. & Bertrand, M. J. M. More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545 (2016).
Patankar, J. V. & Becker, C. Cell death in the gut epithelium and implications for chronic inflammation. Nat. Rev. Gastroenterol. Hepatol. 17, 543–556 (2020).
Sun, L. et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148, 213–227 (2012).
Chen, X. et al. Translocation of mixed lineage kinase domain-like protein to plasma membrane leads to necrotic cell death. Cell Res. 24, 105–121 (2014).
Dondelinger, Y. et al. MLKL compromises plasma membrane integrity by binding to phosphatidylinositol phosphates. Cell Rep. 7, 971–981 (2014).
Zhang, J. et al. MLKL deficiency inhibits DSS-induced colitis independent of intestinal microbiota. Mol. Immunol. 107, 132–141 (2019).
Günther, C. et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 (2011).
Lehle, A. S. et al. Intestinal inflammation and dysregulated immunity in patients with inherited Caspase-8 deficiency. Gastroenterology 156, 275–278 (2019).
Gunther, C. et al. Caspase-8 regulates TNF-α-induced epithelial necroptosis and terminal ileitis. Nature 477, 335–339 (2011).
Loynes, C. A. et al. PGE2 production at sites of tissue injury promotes an anti-inflammatory neutrophil phenotype and determines the outcome of inflammation resolution in vivo. Sci. Adv. 4, eaar8320 (2018).
Duffin, R. et al. Prostaglandin E(2) constrains systemic inflammation through an innate lymphoid cell-IL-22 axis. Science 351, 1333–1338 (2016).
Glas, J. et al. PTGER4 expression-modulating polymorphisms in the 5p13.1 region predispose to Crohn’s disease and affect NF-κB and XBP1 binding sites. PLoS ONE 7, e52873 (2012).
Wu, P.-B. et al. Association between PTGER4 polymorphisms and inflammatory bowel disease risk in Caucasian: a meta-analysis. Medicine 99, e19756 (2020).
Libioulle, C. et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 3, e58 (2007).
Kutmon, M. et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 44, D488–D494 (2016).
Martens, M. et al. WikiPathways: connecting communities. Nucleic Acids Res. 49, D613–D621 (2021).
Alikhani, Z. et al. Advanced glycation end products enhance expression of pro-apoptotic genes and stimulate fibroblast apoptosis through cytoplasmic and mitochondrial pathways. J. Biol. Chem. 280, 12087–12095 (2005).
Ciccocioppo, R. et al. Role of the advanced glycation end products receptor in Crohn’s disease inflammation. World J. Gastroenterol. 19, 8269–8281 (2013).
Hu, W. et al. Notch3 pathway alterations in ovarian cancer. Cancer Res. 74, 3282–3293 (2014).
Anany, M. A. et al. Soluble TNF-like weak inducer of apoptosis (TWEAK) enhances poly(I:C)-induced RIPK1-mediated necroptosis. Cell Death Dis. 9, 1084 (2018).
Bhattacharjee, M. et al. A bioinformatics resource for TWEAK–Fn14 signaling pathway. J. Signal Transduct. 2012, 376470 (2012).
Chopra, M. et al. Blocking TWEAK-Fn14 interaction inhibits hematopoietic stem cell transplantation-induced intestinal cell death and reduces GVHD. Blood 126, 437–444 (2015).
Raaijmakers, L. M. et al. PhosphoPath: visualization of phosphosite-centric dynamics in temporal molecular networks. J. Proteome Res. 14, 4332–4341 (2015).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
He, S. et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-α. Cell 137, 1100–1111 (2009).
Moriwaki, K., Bertin, J., Gough, P. J., Orlowski, G. M. & Chan, F. K. Differential roles of RIPK1 and RIPK3 in TNF-induced necroptosis and chemotherapeutic agent-induced cell death. Cell Death Dis. 6, e1636 (2015).
Samson, A. L. et al. A toolbox for imaging RIPK1, RIPK3, and MLKL in mouse and human cells. Cell Death Differ. https://doi.org/10.1038/s41418-021-00742-x (2021).
Xu, D. et al. TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491 (2018).
Liu, S. et al. MLKL forms disulfide bond-dependent amyloid-like polymers to induce necroptosis. Proc. Natl Acad. Sci. USA 114, E7450–E7459 (2017).
Guo, X. et al. TAK1 regulates caspase 8 activation and necroptotic signaling via multiple cell death checkpoints. Cell Death Dis. 7, e2381 (2016).
Geng, J. et al. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat. Commun. 8, 359 (2017).
Loynes, C. A. et al. PGE2 production at sites of tissue injury promotes an anti-inflammatory neutrophil phenotype and determines the outcome of inflammation resolution in vivo. Sci. Adv. 4, eaar8320 (2018).
Watanabe, Y. et al. KAG-308, a newly-identified EP4-selective agonist shows efficacy for treating ulcerative colitis and can bring about lower risk of colorectal carcinogenesis by oral administration. Eur. J. Pharmacol. 754, 179–189 (2015).
Kabashima, K. et al. The prostaglandin receptor EP4 suppresses colitis, mucosal damage and CD4 cell activation in the gut. J. Clin. Invest. 109, 883–893 (2002).
Sheng, H., Shao, J., Morrow, J. D., Beauchamp, R. D. & DuBois, R. N. Modulation of apoptosis and Bcl-2 expression by prostaglandin E2 in human colon cancer cells. Cancer Res. 58, 362–366 (1998).
Matsumoto, Y. et al. Epithelial EP4 plays an essential role in maintaining homeostasis in colon. Sci. Rep. 9, 15244 (2019).
Vaira, S. et al. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J. Clin. Invest. 118, 2088–2097 (2008).
Gullo, C. et al. Inhibition of proliferation and induction of apoptosis in multiple myeloma cell lines by CD137 ligand signaling. PLoS ONE 5, e10845 (2010).
Kwajah, M. M. S., Mustafa, N., Holme, A. L., Pervaiz, S. & Schwarz, H. Biphasic activity of CD137 ligand-stimulated monocytes on T cell apoptosis and proliferation. J. Leukoc. Biol. 89, 707–720 (2011).
Kobayashi, Y. et al. Prostaglandin E2 enhances osteoclastic differentiation of precursor cells through protein kinase A-dependent phosphorylation of TAK1. J. Biol. Chem. 280, 11395–11403 (2005).
Simmons, A. N., Kajino-Sakamoto, R. & Ninomiya-Tsuji, J. TAK1 regulates Paneth cell integrity partly through blocking necroptosis. Cell Death Dis. 7, e2196 (2016).
Orning, P. et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362, 1064–1069 (2018).
Knosel, T., Schewe, C., Petersen, N., Dietel, M. & Petersen, I. Prevalence of infectious pathogens in Crohn’s disease. Pathol. Res Pr. 205, 223–230 (2009).
Hangai, S. et al. PGE2 induced in and released by dying cells functions as an inhibitory DAMP. Proc. Natl Acad. Sci. USA 113, 3844–3849 (2016).
Rauch, I. et al. NAIP–NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity 46, 649–659 (2017).
Nakase, H. et al. Effect of EP4 agonist (ONO-4819CD) for patients with mild to moderate ulcerative colitis refractory to 5-aminosalicylates: a randomized phase II, placebo-controlled trial. Inflamm. Bowel Dis. 16, 731–733 (2010).
Markovic, T., Jakopin, Z., Dolenc, M. S. & Mlinaric-Rascan, I. Structural features of subtype-selective EP receptor modulators. Drug Disco. Today 22, 57–71 (2017).
Toyoda, Y. et al. Ligand binding to human prostaglandin E receptor EP4 at the lipid-bilayer interface. Nat. Chem. Biol. 15, 18–26 (2019).
Gunther, C. et al. The pseudokinase MLKL mediates programmed hepatocellular necrosis independently of RIPK3 during hepatitis. J. Clin. Invest. 126, 4346–4360 (2016).
Billot, X. et al. Discovery of a potent and selective agonist of the prostaglandin EP4 receptor. Bioorg. Med. Chem. Lett. 13, 1129–1132 (2003).
Young, R. N. et al. Discovery and synthesis of a potent, selective and orally bioavailable EP4 receptor agonist. Heterocycles 64, 437–446 (2004).
Arns, S., Gibe, R., Moreau, A., Monzur Morshed, M. & Young, R. N. Design and synthesis of novel bone-targeting dual-action pro-drugs for the treatment and reversal of osteoporosis. Bioorg. Med. Chem. 20, 2131–2140 (2012).
Billot, X., Colucci, J., Han, Y., Wilson, M.-C. & Young, R. N. EP4 receptor agonist, compositions and methods thereof. Google Patents US7238710B2, 2007.
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Noble, C. L. et al. Regional variation in gene expression in the healthy colon is dysregulated in ulcerative colitis. Gut 57, 1398–1405 (2008).
Haberman, Y. et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J. Clin. Invest. 124, 3617–3633 (2014).
Haberman, Y. et al. Ulcerative colitis mucosal transcriptomes reveal mitochondriopathy and personalized mechanisms underlying disease severity and treatment response. Nat. Commun. 10, 38 (2019).
Fenton, C. G., Taman, H., Florholmen, J., Sorbye, S. W. & Paulssen, R. H. Transcriptional signatures that define ulcerative colitis in remission. Inflamm. Bowel Dis. 27, 94–105 (2021).
Arijs, I. et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 58, 1612–1619 (2009).
Vanhove, W. et al. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm. Bowel Dis. 21, 2673–2682 (2015).
Li, Y. et al. COX-2–PGE2 signaling impairs intestinal epithelial regeneration and associates with TNF inhibitor responsiveness in ulcerative colitis. EBioMedicine 36, 497–507 (2018).
Scheibe, K. et al. Inhibiting interleukin 36 receptor signaling reduces fibrosis in mice with chronic intestinal inflammation. Gastroenterology 156, 1082–1097 (2019).
Stark, C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006).
Hornbeck, P. V. et al. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015).
Zundler, S. et al. Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation. Nat. Immunol. 20, 288–300 (2019).
Acknowledgements
Funding: DFG projects SFB1181 (C02, C05), TRR241 (A02, A03, A08, B04, B05, C02, C04 and INF: IBDome), KFO257 (TP01) and individual grant BE3686/2. Further support: Interdisciplinary Center for Clinical Research (IZKF: J68, A76). S.K. and R.N.Y. received funding from Crohn’s and Colitis Canada. J.V.P. received a post-doctoral fellowship from the Alexander von Humboldt foundation. We thank S. Reid from the Sonnewald laboratory for help with array scanning.
Author information
Authors and Affiliations
Contributions
Study concept, literature search, experimentation, analysis, interpretation of data and critical revision of manuscript: J.V.P., R.N.Y. and C.B. Experimentation and analysis: J.P., T.M.M., S.K., M.G.A., F.M., K.S., M.M., C.H., Y.Y., W.L., M.J.M. and S.Z. Material support: K.J., B.R,. C.G., M.L., S.W., C.N. and A.A.K. Manuscript drafting: J.V.P. and C.B. Intellectual contributions, manuscript editing and material support: A.A.K., B.R., M.F.N., K.J., R.A. and C.B.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Shigekazu Nagata, Arthur Kaser and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
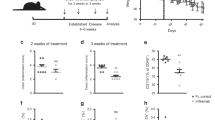
Extended data Fig. 1 EP4 expression is associated with improved IBD outcome.
a, Expression levels (normalized counts) of PTGER4 from the indicated publicly available RNA-Seq studies from IBD patient gut tissues. b, mRNA expression levels (log2 fold change) of the inflammation marker S100A9 by qPCR at various inflammation grades (scores) of intestinal tissues derived from patients with Crohn’s disease (CD) and ulcerative colitis (UC). (n, CD = score 0 = 32; 1 = 19; 2 = 21; 3 = 18) (n, UC = score 0 = 15; 1 = 11; 2 = 20; 3 = 16, geometric mean ± geometric SD). c, Flare-free survival in CD patients with a high versus a low expression levels of PTGER4 d, change in swelling response of mouse small intestinal organoids induced by the EP4 selective agonist, EP4D, upon treatment with increasing concentrations of either LCL161,982 (EP4 receptor antagonist) or TG6–10–1 (EP2 receptor antagonist).
Extended data Fig. 2 Transcriptional signature of PGE2 synthesis is associated with resolution of colitis.
a, KEGG pathway gene ontologies (-log10 p values) of the up- and down-regulated genes in C57BL/6 mice challenged with DSS and receiving EP4D versus those that received vehicle. b, KEGG pathway gene ontologies (-log10 p values) of the up and down-regulated genes in C57BL/6 mice at the high inflammation (Inf_High) and fully resolved (Res_Ful) time points of DSS-induced colitis. c, KEGG pathway gene ontologies (-log10 p values) of the up and down-regulated genes in ulcerative colitis patients from the indicated publicly available dataset. Bars for the pathways of arachidonic acid and linoleic acid metabolism are hashed.
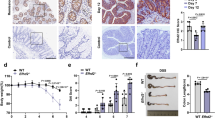
Extended data Fig. 3 IEC-specific ablation of Casp8 increases non-apoptotic death.
a, TUNEL positive cells per 10 crypts, dots represent biological replicates of C57BL/6 mice that were challenged with DSS and received either Vehicle or two different doses of EP4-D (n = 10, mean ± SD, *** = p < 0.001, one way ANOVA, followed by Dunnett’s multiple comparisons test). b, representative micrographs of colonic tissue sections immunostained for phosphorylated MLKL from C57BL/6 mice that were challenged with DSS and received either Vehicle or EP4-D. c, body weight excursions in Casp8fl/fl and Casp8ΔIEC mice challenged with the indicated percentages of DSS. (scale bars = 250 µm) d, survival of Casp8fl/fl and Casp8ΔIEC mice challenged with 1.5% DSS. e, representative micrographs of colonic tissue sections from Casp8ΔIEC and Casp8ΔIEC X Mlkl-/- mice co-stained for phosphorylated MLKL and TUNEL. (scale bars = 100 µm) f, representative micrographs of colonic tissue sections from Casp8fl/fl and Casp8ΔIEC mice challenged with DSS immunostained for caspase 8. (scale bars = 150 µm) g, tamoxifen treatment scheme to induce the deletion of Casp8 specifically in the IEC, followed by DSS a challenge and body weight excursions in the inducible Casp8iΔIEC mice, challenged either with vehicle (Veh) or with EP4-D (n = 3, mean ± SD, at day 4 p = 0.04, at day 5 p = 0.01, two way ANOVA, followed by Sidak’s multiple comparisons test).
Extended data Fig. 4 Selective EP4 activation suppresses Casp8-independent, TNF-induced IEC death.
a, intestinal organoids from Casp8fl/fl and Casp8ΔIEC mice treated as indicated and stained with PI to reveal dead cells followed by quantification of PI intensity (n = 4, mean ± SD, *** = p 0.0002, one way ANOVA, followed by Tukey’s multiple comparisons). b, intestinal organoids from Casp8ΔIEC mice treated as indicated and stained with PI to reveal dead cells followed by quantification of PI intensity (n = 4, mean ± SD, *** = p < 0.001 compared versus TNF, one way ANOVA, followed by Dunnett’s multiple comparisons). c, gating scheme for HT-29 cells subjected to cell death treatments followed by flow cytometric evaluation. d, densitometric quantification of the blots for pRIPK1 from Fig. 5h (n = 3, mean ± SD, ** = p 0.003, *** = p 0.0003, compared versus TSz, one way ANOVA, followed by Dunnett’s multiple comparisons).
Extended data Fig. 5 EP4 is the primary E-type prostanoid receptor on IEC.
a, Micrographs of PI and caspase 3/7 activity probe stained, biopsy-derived colonic organoids from human volunteers treated as indicated. b, normalized counts (fpkm) of Ptger2 and Ptger4 from C57BL/6 mouse intestinal organoids subjected to RNA-Sequencing and normalized counts (log2 TPM) from the indicated publicly available single-cell RNA sequencing dataset from the mouse small intestine.
Supplementary information
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots for Fig. 5
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 7
Statistical Source Data
Source Data Fig. 7
Unprocessed Western Blots for Fig. 7
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Rights and permissions
About this article
Cite this article
Patankar, J.V., Müller, T.M., Kantham, S. et al. E-type prostanoid receptor 4 drives resolution of intestinal inflammation by blocking epithelial necroptosis. Nat Cell Biol 23, 796–807 (2021). https://doi.org/10.1038/s41556-021-00708-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00708-8
This article is cited by
-
EFHD2 suppresses intestinal inflammation by blocking intestinal epithelial cell TNFR1 internalization and cell death
Nature Communications (2024)
-
Prostaglandin E2 controls the metabolic adaptation of T cells to the intestinal microenvironment
Nature Communications (2024)
-
Formyl peptide receptor 2 as a potential therapeutic target for inflammatory bowel disease
Acta Pharmacologica Sinica (2023)
-
Endothelial Caspase-8 prevents fatal necroptotic hemorrhage caused by commensal bacteria
Cell Death & Differentiation (2023)
-
Apoptotic cell death in disease—Current understanding of the NCCD 2023
Cell Death & Differentiation (2023)