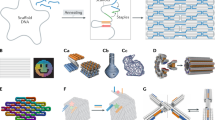
Abstract
Recently, DNA has been used to make nanodevices for a myriad of applications across fields including medicine, nanomanufacturing, synthetic biology, biosensing and biophysics. However, current DNA nanodevices rely primarily on geometric design, and it remains challenging to rationally design functional properties such as force-response or actuation behaviour. Here we report an iterative design pipeline for DNA assemblies that integrates computer-aided engineering based on coarse-grained molecular dynamics with a versatile computer-aided design approach that combines top-down automation with bottom-up control over geometry. This intuitive framework allows for rapid construction of large, multicomponent assemblies from three-dimensional models with finer control over the geometrical, mechanical and dynamical properties of the DNA structures in an automated manner. This approach expands the scope of structural complexity and enhances mechanical and dynamic design of DNA assemblies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Original data for TEM images and gel electrophoresis are included as Source data. The remaining data supporting the findings of this study are available within the Article and its supplementary information files or available from the corresponding author upon reasonable request.
Code availability
The developed design software MagicDNA is available from GitHub at https://github.com/cmhuang2011/MagicDNA.
References
Chang, K.-H. e-Design: Computer-Aided Engineering Design (Academic, 2016).
Olson, G. B. Computational design of hierarchically structured materials. Science 277, 1237–1242 (1997).
Panchal, J. H., Kalidindi, S. R. & McDowell, D. L. Key computational modeling issues in integrated computational materials engineering. Comput. Aided Des. 45, 4–25 (2013).
Backman, D. G. et al. ICME at GE: accelerating the insertion of new materials and processes. JOM 58, 36–41 (2006).
Huang, P.-S., Boyken, S. E. & Baker, D. The coming of age of de novo protein design. Nature 537, 320–327 (2016).
Douglas, S. M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Williams, S. et al. in DNA Computing (eds Goel, A., Simmel, F. C. & Sosík, P.) 90–101 (Springer, 2008).
Benson, E. et al. DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015).
Veneziano, R. et al. Designer nanoscale DNA assemblies programmed from the top down. Science 352, aaf4388 (2016).
Doye, J. P. K. et al. Coarse-graining DNA for simulations of DNA nanotechnology. Phys. Chem. Chem. Phys. 15, 20395–20414 (2013).
Snodin, B. E. K. et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 142, 234901 (2015).
Maffeo, C. & Aksimentiev, A. MrDNA: a multi-resolution model for predicting the structure and dynamics of DNA systems. Nucleic Acids Res. 48, 5135–5146 (2020).
Reshetnikov, R. V. et al. A coarse-grained model for DNA origami. Nucleic Acids Res. 46, 1102–1112 (2018).
Seeman, N. C. Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982).
Rothemund, P. W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Jiang, Q. et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 134, 13396–13403 (2012).
Sun, W. et al. Casting inorganic structures with DNA molds. Science 346, 1258361 (2014).
Liu, X. et al. Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018).
Shaw, A. et al. Spatial control of membrane receptor function using ligand nanocalipers. Nat. Methods 11, 841–846 (2014).
Shen, B. et al. Plasmonic nanostructures through DNA-assisted lithography. Sci. Adv. 4, eaap8978 (2018).
Le, J. V. et al. Probing nucleosome stability with a DNA origami nanocaliper. ACS Nano 10, 7073–7084 (2016).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Marras, A. E., Zhou, L., Su, H.-J. & Castro, C. E. Programmable motion of DNA origami mechanisms. Proc. Natl Acad. Sci. USA 112, 713–718 (2015).
Zhou, L., Marras, A. E., Huang, C.-M., Castro, C. E. & Su, H.-J. Paper origami-inspired design and actuation of DNA nanomachines with complex motions. Small 14, 1802580 (2018).
Jun, H. et al. Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 13, 2083–2093 (2019).
Jun, H. et al. Autonomously designed free-form 2D DNA origami. Sci. Adv. 5, eaav0655 (2019).
Jun, H., Wang, X., Bricker, W. P. & Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 10, 5419 (2019).
Zhou, L., Marras, A. E., Su, H.-J. & Castro, C. E. DNA origami compliant nanostructures with tunable mechanical properties. ACS Nano 8, 27–34 (2014).
Ke, Y. et al. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 131, 15903–15908 (2009).
Pandey, S. et al. Algorithmic design of self-folding polyhedra. Proc. Natl Acad. Sci. USA 108, 19885–19890 (2011).
Castro, C. E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Kim, D.-N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012).
Poppleton, E. et al. Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation. Nucleic Acids Res. 48, e72 (2020).
Snodin, B. E. K., Schreck, J. S., Romano, F., Louis, A. A. & Doye, J. P. K. Coarse-grained modelling of the structural properties of DNA origami. Nucleic Acids Res. 47, 1585–1597 (2019).
Shi, Z., Castro, C. E. & Arya, G. Conformational dynamics of mechanically compliant DNA nanostructures from coarse-grained molecular dynamics simulations. ACS Nano 11, 4617–4630 (2017).
Sharma, R., Schreck, J. S., Romano, F., Louis, A. A. & Doye, J. P. K. Characterizing the motion of jointed DNA nanostructures using a coarse-grained model. ACS Nano 11, 12426–12435 (2017).
Huang, C.-M., Kucinic, A., Le, J. V., Castro, C. E. & Su, H.-J. Uncertainty quantification of a DNA origami mechanism using a coarse-grained model and kinematic variance analysis. Nanoscale 11, 1647–1660 (2019).
Engelhardt, F. A. S. et al. Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano 13, 5015–5027 (2019).
Wei, R., Martin, T. G., Rant, U. & Dietz, H. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 51, 4864–4867 (2012).
Johnson, J. A., Dehankar, A., Winter, J. O. & Castro, C. E. Reciprocal control of hierarchical DNA origami–nanoparticle assemblies. Nano Lett. 19, 8469–8475 (2019).
Ponnuswamy, N. et al. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 8, 15654 (2017).
Dietz, H., Douglas, S. M. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009).
Zhang, F. et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 10, 779–784 (2015).
Bertoldi, K., Vitelli, V., Christensen, J. & van Hecke, M. Flexible mechanical metamaterials. Nat. Rev. Mater. 2, 17066 (2017).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 1–23 (2017).
Wagenbauer, K. F., Sigl, C. & Dietz, H. Gigadalton-scale shape-programmable DNA assemblies. Nature 552, 78–83 (2017).
Tikhomirov, G., Petersen, P. & Qian, L. Fractal assembly of micrometre-scale DNA origami arrays with arbitrary patterns. Nature 552, 67–71 (2017).
Marchi, A. N. et al. Origami. Nano Lett. 14, 5740–5747 (2014).
Ong, L. L. et al. Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components. Nature 552, 72–77 (2017).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Sobczak, J.-P. J., Martin, T. G., Gerling, T. & Dietz, H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science 338, 1458–1461 (2012).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Abramoff, M. D., Magalhães, P. J. & Ram, S. J. Image Processing with ImageJ. Biophotonics International 11, 36–42 (2004).
Acknowledgements
This work was supported by National Science Foundation (NSF) grant 1536862 to H.-J.S. and C.E.C. and grant 1921881 to C.E.C. We acknowledge F. Engelhardt and H. Dietz for providing custom scaffolds, T. Aksel and S. Douglas for sharing the caDNAno toolkit, C. Maffeo and A. Aksimentiev for supporting the interface to MrDNA, T. MacCulloch and N. Stephanopoulos for providing K10 peptide and A. Tran, P. Le and P. Lukeman for testing MagicDNA Runtime packages. We thank the Campus Microscopy and Imaging Facility (CMIF) of The Ohio State University for imaging support. We also thank W. Pfeifer and C. Maffeo for critiques on the manuscript and supplementary material. Funding provided by NSF (NSF CMMI) 1536862 and NSF (NSF CMMI) 1921881.
Author information
Authors and Affiliations
Contributions
C.-M.H. developed the software and the algorithm, designed and simulated all the structures, analysed the data, prepared the tutorial and supported experiments. A.K. conducted the majority of experiments and analysed the experimental results. J.A.J. was an initial user and provided critical early feedback on software features, interface and instructions. H.-J.S. supervised the development of the software and interpreted the data. C.E.C. supervised the experimental validation and the entire study, supported the development of the software and interpreted the data. C.-M.H., A.K., H.-J.S. and C.E.C. wrote the manuscript. All authors commented on and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Materials thanks Ebbe Andersen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–3, Figs. 1–87, Table 1, captions for Videos 1 and 2, and References 1–27.
Supplementary Video 1
Top-down parametric design for a hinge structure performed by converting two lines into bundles and specifying the connectivity to assemble the components.
Supplementary Video 2
The final design profile of the airplane and the CG simulation with 3 × 108 steps.
Supplementary Data 3
The Excel sheets for the staple list of the 14 structures for fabrication.
Supplementary Data 4
Unprocessed gels and gel-intensity analysis for yield determination.
Source data
Source Data Fig. 1
MagicDNA design file and unprocessed TEM image.
Source Data Fig. 2
DNA origami structure raw TEM images.
Source Data Fig. 2d
Source data for motion analysis of 4 bar mechanism.
Source Data Fig. 3
DNA origami structure raw TEM images.
Source Data Fig. 4
DNA origami structure raw TEM images.
Source Data Fig. 6
DNA origami structure raw TEM images and gel electrophoresis image depicting robot arm assembly.
Rights and permissions
About this article
Cite this article
Huang, CM., Kucinic, A., Johnson, J.A. et al. Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 20, 1264–1271 (2021). https://doi.org/10.1038/s41563-021-00978-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-021-00978-5
This article is cited by
-
Mechanism of DNA origami folding elucidated by mesoscopic simulations
Nature Communications (2024)
-
Employing toehold-mediated DNA strand displacement reactions for biomedical applications
Med-X (2024)
-
DNA T-shaped crossover tiles for 2D tessellation and nanoring reconfiguration
Nature Communications (2023)
-
Throwing and manipulating and cheating with a DNA nano-dice
Nature Communications (2023)
-
Fabricating higher-order functional DNA origami structures to reveal biological processes at multiple scales
NPG Asia Materials (2023)