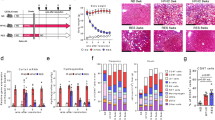
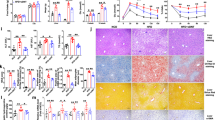
Abstract
Nonalcoholic steatohepatitis (NASH) is a manifestation of systemic metabolic disease related to obesity, and causes liver disease and cancer1,2. The accumulation of metabolites leads to cell stress and inflammation in the liver3, but mechanistic understandings of liver damage in NASH are incomplete. Here, using a preclinical mouse model that displays key features of human NASH (hereafter, NASH mice), we found an indispensable role for T cells in liver immunopathology. We detected the hepatic accumulation of CD8 T cells with phenotypes that combined tissue residency (CXCR6) with effector (granzyme) and exhaustion (PD1) characteristics. Liver CXCR6+ CD8 T cells were characterized by low activity of the FOXO1 transcription factor, and were abundant in NASH mice and in patients with NASH. Mechanistically, IL-15 induced FOXO1 downregulation and CXCR6 upregulation, which together rendered liver-resident CXCR6+ CD8 T cells susceptible to metabolic stimuli (including acetate and extracellular ATP) and collectively triggered auto-aggression. CXCR6+ CD8 T cells from the livers of NASH mice or of patients with NASH had similar transcriptional signatures, and showed auto-aggressive killing of cells in an MHC-class-I-independent fashion after signalling through P2X7 purinergic receptors. This killing by auto-aggressive CD8 T cells fundamentally differed from that by antigen-specific cells, which mechanistically distinguishes auto-aggressive and protective T cell immunity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All raw gene RNA-seq data from mice have been deposited in the GEO under accession number GSE145104. Single-cell RNA-seq data of fat tissue from patients with NASH can be found in the under accession number EGAS00001004738, and can be accessed at https://ega-archive.org/access/data-access upon request to the corresponding author. Single-cell RNA-seq data from livers of patients with steatosis or NASH can be found under accession number GSE159977. All datasets are available from the corresponding author upon request. Source data are provided with this paper.
Change history
10 May 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-03568-2
References
Roden, M. & Shulman, G. I. The integrative biology of type 2 diabetes. Nature 576, 51–60 (2019).
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M. & Sanyal, A. J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922 (2018).
Hotamisligil, G. S. Inflammation, metaflammation and immunometabolic disorders. Nature 542, 177–185 (2017).
Wolf, M. J. et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell 26, 549–564 (2014).
Malehmir, M. et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 25, 641–655 (2019).
Clapper, J. R. et al. Diet-induced mouse model of fatty liver disease and nonalcoholic steatohepatitis reflecting clinical disease progression and methods of assessment. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G483–G495 (2013).
Ma, C. et al. NAFLD causes selective CD4+ T lymphocyte loss and promotes hepatocarcinogenesis. Nature 531, 253–257 (2016).
Fernandez-Ruiz, D. et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016).
Topham, D. J. & Reilly, E. C. Tissue-resident memory CD8+ T cells: from phenotype to function. Front. Immunol. 9, 515 (2018).
Böttcher, J. P. et al. Functional classification of memory CD8+ T cells by CX3CR1 expression. Nat. Commun. 6, 8306 (2015).
Gerlach, C. et al. The chemokine receptor CX3CR1 defines three antigen-experienced CD8 T cell subsets with distinct roles in immune surveillance and homeostasis. Immunity 45, 1270–1284 (2016).
Olson, J. A., McDonald-Hyman, C., Jameson, S. C. & Hamilton, S. E. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity 38, 1250–1260 (2013).
Khan, O. et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature 571, 211–218 (2019).
Alfei, F. et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269 (2019).
Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463 (2016).
Veldhoen, M. Interleukin 17 is a chief orchestrator of immunity. Nat. Immunol. 18, 612–621 (2017).
Man, K. & Kallies, A. Synchronizing transcriptional control of T cell metabolism and function. Nat. Rev. Immunol. 15, 574–584 (2015).
Wang, Z. et al. BART: a transcription factor prediction tool with query gene sets or epigenomic profiles. Bioinformatics 34, 2867–2869 (2018).
Leonard, W. J., Lin, J. X. & O’Shea, J. J. The γc family of cytokines: basic biology to therapeutic ramifications. Immunity 50, 832–850 (2019).
Hedrick, S. M., Hess Michelini, R., Doedens, A. L., Goldrath, A. W. & Stone, E. L. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 12, 649–661 (2012).
Cepero-Donates, Y. et al. Interleukin-15-mediated inflammation promotes non-alcoholic fatty liver disease. Cytokine 82, 102–111 (2016).
Chirifu, M. et al. Crystal structure of the IL-15–IL-15Rα complex, a cytokine-receptor unit presented in trans. Nat. Immunol. 8, 1001–1007 (2007).
Balmer, M. L. et al. Memory CD8+ T cells require increased concentrations of acetate induced by stress for optimal function. Immunity 44, 1312–1324 (2016).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074 (2019).
Liu, X. et al. Acetate production from glucose and coupling to mitochondrial metabolism in mammals. Cell 175, 502–513 (2018).
Bachem, A. et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297 (2019).
Voskoboinik, I., Whisstock, J. C. & Trapani, J. A. Perforin and granzymes: function, dysfunction and human pathology. Nat. Rev. Immunol. 15, 388–400 (2015).
Chang, C. H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Linden, J., Koch-Nolte, F. & Dahl, G. Purine release, metabolism, and signaling in the inflammatory response. Annu. Rev. Immunol. 37, 325–347 (2019).
Chekeni, F. B. et al. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature 467, 863–867 (2010).
Stark, R. et al. TRM maintenance is regulated by tissue damage via P2RX7. Sci. Immunol. 3, eaau1022 (2018).
Borges da Silva, H. et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8+ T cells. Nature 559, 264–268 (2018).
Zhao, J. et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 6, 22 (2020).
Setoguchi, R. IL-15 boosts the function and migration of human terminally differentiated CD8+ T cells by inducing a unique gene signature. Int. Immunol. 28, 293–305 (2016).
Lampl, S. et al. Reduced mitochondrial resilience enables non-canonical induction of apoptosis after TNF receptor signaling in virus-infected hepatocytes. J. Hepatol. 73, 1347–1359 (2020).
Chen, H. et al. Cytofkit: a Bioconductor package for an integrated mass cytometry data analysis pipeline. PLoS Comput. Biol. 12, e1005112 (2016).
Mackay, L. K. et al. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015).
Li, M. Z. & Elledge, S. J. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4, 251–256 (2007).
Guillot, A. & Tacke, F. The unexpected role of neutrophils for resolving liver inflammation by transmitting microRNA-223 to macrophages. Hepatology 71, 749–751 (2020).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhou, Q. et al. A mouse tissue transcription factor atlas. Nat. Commun. 8, 15089 (2017).
Dreos, R., Ambrosini, G., Groux, R., Cavin Périer, R. & Bucher, P. The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 45, D51–D55 (2017).
Khan, A. et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266 (2018).
Kulakovskiy, I. V. et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-seq analysis. Nucleic Acids Res. 46, D252–D259 (2018).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Zappia, L. & Oshlack, A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience 7, (2018).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Mahi, N. A., Najafabadi, M. F., Pilarczyk, M., Kouril, M. & Medvedovic, M. GREIN: an interactive web platform for re-analyzing GEO RNA-seq data. Sci. Rep. 9, 7580 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We thank S. Yousuf and M. Qui for technical support in preparing samples for single-cell RNA-seq analysis; P. Sinn for technical support; the personnel (particularly J. Maurer) of the Core Facility Flow Cytometry at Biomedical Center Munich (LMU), for providing the imaging cytometry instrument and analysis software; and the personnel of the Core Facility Cell Sorting of the German Cancer Research Center Heidelberg. This work was supported by the DFG, project number 424926990, to J.P.B., CIBSS EXC-2189 project identifier 390939984 to E.J.P., SFB TRR179 and DZIF Munich site to P.A.K.
Author information
Authors and Affiliations
Contributions
M.D. and P.A.K. designed the study. M.D., D.P., P.F., A.S., M.L., P.M., F.B., D.I., J.W., S.H., M.A., A.G., A.B., D. Heide, F.M. and S. Reider performed experiments. S.D., R.Ö. and A.M.K. undertook bioinformatics analysis. D. Hartmann, N.H., M.S., C.G.-C., T.G., G.S., J.-P.M., M.E., S. Roth, A.T.B., B.M.-S., F.K.-N., R.K., T.B., P.R., V.L., J.-F.D. and M.H. provided tissue samples, essential reagents or mice. R.R., E.J.P., H.T., F.T., J.-F.D., D. Haller, P.J.M., R.H., R.T., D.Z., J.P.B., M.H. and P.A.K. contributed scientific input. M.D. and P.A.K. wrote the manuscript, and all authors contributed to feedback and proofreading.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Paul Klenerman, Ignacio Melero, Daniel Peeper and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Immune-cell characteristics in livers of mice fed a CD-HFD.
a–e, Liver damage after 12 months of feeding a CD-HFD (which induces features of NASH) in Ja18−/− and Cd1d−/− mice, determined by sALT and NAS with representative images of H&E staining (mice fed a normal diet, n = 8; CD-HFD-fed, wild-type mice, n ≥ 7; CD-HFD-fed, Ja18−/− mice, n ≥ 7; CD-HFD-fed, Cd1d−/− mice: n ≥ 8). Scale bar, 100 μm. Two independent experiments. f, Body weight of mice over the course of six months. g, h, Numbers of hepatic lymphocytes (g) and hepatic CXCR6+ T cell populations (h) in mice fed a normal diet (n = 11) or CD-HFD (n = 13). Two independent experiments.i–k, Numbers and phenotype of hepatic CD44+ CXCR6+ and CXCR6− CD8 T cells in NASH mice (n ≥ 5). Two independent experiments. l, Frequencies of CXCR6+GzmB+ CD8 T cells in different organs in mice fed a normal diet (n = 4) and NASH mice (n = 4). m–o, Localization and quantification of hepatic CXCR6+PD1high CD8 T cells in mice fed a normal diet (n = 3) and NASH mice (n = 3), by confocal microscopy with representative images. Scale bar, 200 μm. CD3 (yellow), CD8 (purple), CXCR6 (cyan), PD1 (red) and collagen IV (white). Rectangles highlighted in m indicate representative zoomed areas shown in Fig. 1g. ns, not significant; nd, not determined. Exact P (a–d, f–l, n, o) and n (a–d, f, i–k) values are presented in Source Data. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA with Sidak’s (g, h, l) or with Tukey’s multiple comparison test (j, k), one-way ANOVA with Tukey’s (a–d) or with Dunnett’s multiple comparison test (f) and unpaired two-tailed t-test (i, n, o). In a–d, f–l, n, o, data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 2 Increased numbers of hepatic CXCR6+PD1high CD8 T cells in different mouse models of obesity-related NASH.
a, b, Confocal microscopy of hepatic PD1high CD8 T cells and IMARIS-based quantification with representative images (normal diet, n = 5; high-fat diet (HFD), n = 4; CD-HFD, n = 6). Scale bars, 500 μm (left), 100 μm (right). CD8 (red), PD1 (green), CK19 (blue), IBA1 (cyan), αSMA (purple) and DAPI (white). Rectangles highlighted in the left panel indicate represent zoomed areas in the right panel. Red arrows indicate PD1− CD8 T cells. Yellow arrows indicate PD1high CD8 T cells. One-way ANOVA with Dunnett’s multiple comparison test. c, d, Correlation of liver damage (NAS) with frequencies of hepatic PD1high CD8 T cells from mice fed a normal diet or CD-HFD. Coefficient of determination (R2) and statistical significance (P value) were determined using Pearson’s correlation. e, f, CXCR6 and PD1 expression levels in hepatic CD8 T cells in mice fed CD-HFD (e) or a high trans-fat Western diet (WD) (f) (n = 5) for 12 months compared to mice fed a normal diet (n = 5). h, i, Liver damage (sALT and NAS) in mice from f. j, k, CXCR6 and PD1 expression levels in hepatic CD8 T cells in ob/− and ob/ob mice after 4 months of chow feeding or mice fed a normal diet (ob/−, n = 6; ob/ob, n = 4). l, m, Liver damage (sALT and NAS) in mice from j. Exact P values (b, f–m) are presented in Source Data. *P < 0.05, **P < 0.01, ****P < 0.0001. Unpaired two-tailed t-test (f–m). In b, f–m, data are mean ± s.e.m., errors are shown as s.d.
Extended Data Fig. 3 Transcriptome analysis of hepatic CXCR6+ CD8 T cells from NASH mice.
a, Volcano plot showing differentially expressed genes between CXCR6+ CD8 T cells from NASH mice (fed a CD-HFD) compared to healthy mice (fed a normal diet). b, GSEA for distinct pathways from a. c, Flow cytometry analysis of protein levels of transcription factors in hepatic CXCR6+ and CXCR6− CD8 T cells from healthy and NASH mice (n ≥ 5). d, Binding analysis for regulation of transcription (BART) to predict transcription-factor activity in hepatic CXCR6+ compared to CXCR6− CD8 T cells in NASH mice. e, Transcription-factor network analysis created from differentially expressed genes (a) and BART. f, GSEA for transcription-factor-dependent genes from a. g–i, Flow cytometry analysis of FOXO1 expression in hepatic CXCR6+ and CXCR6− CD8 T cells (g, h) or CD4 T cells (i) from mice fed a normal diet and NASH mice (n ≥ 6). j, k, Image-stream analysis of expression and localization of FOXO1 in different T cell populations from spleen and liver of wild-type mice with representative images. Tn, naive T cells; Tcm, central memory T cells; Tem, effector memory T cells. l, Similarity score to determine nuclear localization of FOXO1 and surface localization of CD44 (n = 5). m, n, FOXO1 expression in hepatic CD44+ CD8 T cells in mice fed a Western diet (n = 5) and ob/− or ob/ob mice (n ≥ 4). PI, propidium iodide. Exact P and n values (c, g-i) are presented in Source Data. *P < 0.05, **P < 0.01, ****P < 0.0001. One-way ANOVA (k, l), two-way ANOVA with Tukey’s multiple comparison test (c, i) and unpaired two-tailed t-test (h, m, n). In c, h, i, k–n, data are mean ± s.e.m., errors are shown as s.d.
Extended Data Fig. 4 Hepatic CXCR6+FOXO1low CD8 T cell frequency correlates with liver damage in NASH mice and patients with NASH.
a–c, Hepatic CXCR6+FOXO1low CD8 T cells at 6, 9 and 12 months of feeding a CD-HFD, and correlation of FOXO1 expression levels in hepatic CD44+CD8 T cells with frequency of hepatic CXCR6+PD1high of CD44+ CD8 T cells from mice fed a CD-HFD, and with NAS. Four independent experiments. d, e, CXCR6 and CX3CR1 expression levels in sorted hepatic CD44+CXCR6− CD8 T cells from healthy mice fed a normal diet (n = 5) after 18 h of treatment with the FOXO1 inhibitor AS1842856. f, CXCR6 expression levels after FOXO1 overexpression (transduction with pMP71-Foxo1-eGFP or pMP71-eGFP) in sorted CXCR6+ CD8 T cells (n = 3). g, h, Correlation of sALT and NAS with FOXO1 expression in hepatic CD44+ CD8 T cells from b. i, Correlation of FOXO1 expression in liver CD69+ CD8 T cells with sALT in patients with NASH (n = 15 in total: n = 12 NASH; n = 3 normal liver tissue (NAS = 0)). j, CXCR6 expression quantified by qPCR with reverse transcription (RT–qPCR) in liver tissue of patients with NASH (NAS ≥ 5) (unaffected tissue, n = 6; tissue from patients with NASH, n = 7). k, l, Correlation of relative CXCR6 expression with sALT and NAS in patients with NASH (unaffected individuals, n = 6; patients with NASH, n = 10). m, Correlation of sALT with numbers of cells that are positive for CXCR6 per mm2 of liver tissue in patients with NASH (n = 18). Scale bar, 100 μm. Representative image of in situ hybridization for CXCR6 mRNA in liver of patients with low or high sALT. Exact P (e, f, j) and n (b, c, g, h) values are presented in Source Data. *P < 0.05, **P < 0.01, ****P < 0.0001. Coefficient of determination (R2) and statistical significance (P value) were determined using Pearson’s correlation (b, c, g–i, k–m). Unpaired two-tailed (f, j) and paired two-tailed t-test (e). In e, f, j, data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 5 Increased levels of IL-15 in serum and liver tissue of NASH mice correlate with liver damage and numbers of hepatic CXCR6+ CD8 T cells.
a, Heat map of fold changes for cytokine and chemokine serum concentrations in NASH (fed a CD-HFD) (n ≥ 8) compared to healthy (fed a normal diet) mice (n ≥ 6) after 12 months of feeding. Values above 2.5 are red. Asterisk denotes values that are significantly different. Exact P and n values are presented in Source Data. Two independent experiments. b, Cytokine and chemokine expression quantified by RT–qPCR in the liver tissue of mice fed a normal diet (n = 5) and NASH mice (n = 9). c, Flow cytometry analysis of the increase in CXCR6 expression after incubation of sorted hepatic CD44+CXCR6− CD8 T cells with cytokines for 24 h (n = 13). d, Flow cytometry analysis of FOXO1 expression 24 h after IL-15 stimulation of hepatic CD44+ CD8 T cells (n = 10). e–g, Flow cytometry analysis of FOXO1 expression in human CD8 T cells at 5 d after IL-15 stimulation (15 ng ml−1) and frequencies of CXCR6+CD69+ cells (n = 5). h, PD1, GzmB and CXCR3 expression by mouse hepatic CXCR6+ CD8 T cells 24 h after IL-15 stimulation (n = 9). i, Percentage of CXCR6-expressing CD8 T cells in sorted, splenic CD44+ CD8 T cells 2 d after stimulation with IL-15 (10 ng ml−1) and/or IL-21 (10 ng ml−1) (n = 5). Two independent experiments. j, k, IL-15 serum concentrations in ob/− (n = 4) and hyperphagic, leptin-deficient ob/ob mice (n = 4) fed a chow diet for 4 months, in mice fed a normal diet (n = 4) or a high trans-fat Western diet for 12 months (n = 5). l–n, Correlation of IL-15 serum levels with sALT, NAS and FOXO1 expression in hepatic CD44+ CD8 T cells of NASH mice (6 months, n = 4; 9 months, n = 7; 12 months, n = 13). Four independent experiments. o–q, Correlation of relative hepatic IL-15 gene expression levels with sALT, NAS and CXCR6 gene expression in patients with NASH and in individuals with normal liver (normal, n = 6; NASH, n = 10). Exact P (a–d, f–k) and n (a) values are presented in Source Data. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Coefficient of determination (R2) and statistical significance (P value) was determined using Pearson´s correlation (l–q). One-way ANOVA with Tukey’s (i) or Dunnett’s multiple comparison test (c), and unpaired two-tailed t-test (a, b, d, f–h, j, k). In a–d, f–k, data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 6 Single-cell RNA-seq of CD3+ T cells from livers of patients with steatosis or NASH.
a, UMAP plot visualization of hepatic CD3+ T cell clusters detected in livers from patients with steatosis (n = 4) or NASH (NAS ≥ 5) (n = 3). b, UMAP plot visualization of hepatic CD3+ T cell clusters and of selected genes detected in patients with NASH (NAS ≥ 5) (n = 3). c, Frequencies of CXCR6+ and PDCD1+ cells in CD8 T cells in the RGS1+, PLCG2+ and ITGA1+ cluster in livers from patients with steatosis or NASH. d, Differentially expressed genes in the CD8 RGS1+ cluster between livers of patients with steatosis or NASH. e, Flow cytometry analysis of FOXO1 expression in CD69+ CD8 T cells from livers of unaffected individuals (n = 3) and livers of patients with NASH (n = 5). P = 0.0088, two-tailed t-test, data are mean ± s.e.m. f, UMAP plot visualization of hepatic CD3+ T cell clusters detected in patients with NASH from fat tissue (n = 2). g, Jaccard similarity score between tissue-resident CD8 T cell populations in fat and liver of patients with NASH (n = 2). h, Correlation of mouse RNA-seq dataset from CXCR6+ compared to CXCR6− CD8 T cells in mice fed a CD-HFD with human single-cell RNA-seq dataset from the CD8 GNLY+FGFBP2+ cluster. The solid line is the linear regression, and the grey area shows the 95% confidence intervals. Coefficient of determination (R2) and statistical significance (P value) were determined using Spearman’s correlation.
Extended Data Fig. 7 Anti-CD122 application in NASH mice leads to depletion of CXCR6+GzmB+ CD8 T cells and amelioration of liver damage.
a, CD122 expression on splenic CD44+ CD8 T cells after stimulation with IL-15 (10 ng ml−1) for 24 h (n = 5). b, CD122 expression on hepatic CXCR6+ CD8 T cells (n = 8). Two independent experiments. c–e, Frequencies and cell numbers of CXCR6+PD1+ and CXCR6+GzmB+ CD8 T cells after anti-CD122-treatment for 2 weeks in NASH mice (fed CD-HFD for 12 months) (CD-HFD + immunoglobulin (Ig), n = 6; CD-HFD + anti-CD122, n = 6). f, sALT in NASH mice from e before and after anti-CD122 treatment for 2 weeks. Unpaired two-tailed t-test of sALT values between groups before treatment, P = 0.7137. g–k, Numbers of CX3CR1 CD8 T cells, CD44+ CD4 T cells, natural killer T cells and natural killer (NK) cells in the liver after anti-CD122 antibody application from mice in e. l, m, No change in phenotype of natural killer cells in NASH mice. Dot plots are representative of ≥5 mice. Exact P values (a, b, e–m) are presented in Source Data. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. One-way ANOVA with Tukey’s multiple comparison test (e, g–k) and paired (a, f) and unpaired (b, l, m) two-tailed t-test. In a, b, e–m, data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 8 IL-15-treated CXCR6+ CD8 T cells become auto-aggressive after exposure to acetate.
a, Auto-aggressive activity of IL-15-treated CXCR6+FOXO1low CD8 T cells against hepatocytes (n = 3). b, GzmB-expression levels in CXCR6+ CD8 T cells exposed for 24 h to different concentrations of acetate in vitro (n = 4). c–e, Fluorospot assay with representative images to quantify TNF and IFNγ release from IL-15-stimulated CXCR6− and CXCR6+ CD8 T cells after acetate exposure and from peptide-specific stimulation of OT1 CD8 T cells (n = 5). f, GSEA for differentially expressed genes from hepatic CXCR6+ and CXCR6− CD8 T cells of mice fed a CD-HFD compared to in vitro-generated memory CD8 T cells exposed to acetate or butyrate. g, Acetate concentrations in supernatants of primary mouse hepatocytes after exposure to palmitate for 24 h (250 μM) or with an agonistic trimeric FasL (25 ng ml−1) to induce cell death (n = 6). h, Cytotoxic activity of acetate-exposed polyclonal CD8 T cells after FOXO1 inhibition (AS1842856, 100 nM) against hepatocytes (n = 3) and quantification. i, GzmB expression in acetate-exposed, GFP-expressing CD8 T cells after retroviral transduction with pMP71-GFP-Foxo1 or pMP71-GFP (n = 3). j, k, sALT at day 2 after adoptive transfer of IL-15-stimulated or FOXO1-inhibited and acetate-exposed CD8 T cells (n ≥ 3). l, Calculation of liver-damage-inducing potential per cell for auto-aggressive CD8 T cells (n = 8). m, Surface LAMP1 expression on CXCR6+ CD8 T cells after a 24-h exposure to acetate (15 mM) (n = 9). n, o, Liver damage quantified by NAS and sALT in Prf1−/− and littermate control mice after CD-HFD feeding for 1 year. p, ICAM1 expression on hepatocytes in coculture with IL-15-stimulated, acetate-exposed CXCR6+ CD8 T cells (n = 7). q, Liver immunohistochemistry with representative images and quantification of ICAM1 clusters per mm2 of liver tissue in mice fed a normal diet (n = 5), NASH mice (n = 6) and NASH mice treated with anti-CD122 antibody (n = 6). Scale bar, 100 μm. r, Representative microscopic images of IL-15-stimulated, acetate-exposed CXCR6+ CD8 T cells in contact with hepatocytes and quantification of the distance. Arrows indicate polarized LFA1 orientation on T cell interacting with hepatocyte. Scale bars, 30 μm (left), 15 μm (right). s, Auto-aggressive activity of IL-15-treated CXCR6+ CD8 T cells against hepatocytes in presence of acetate (15 mM) or TNF (5 ng ml−1) and anti-LFA1 antibodies (n = 3). t, sALT at day 2 after adoptive transfer of auto-aggressive CD8 T cells and injection of anti-LFA1 (100 μg per mouse) or control immunoglobulin. Two independent experiments. u, v, TNF expression in livers from unaffected individuals (n = 6) or patients with NASH (n = 7) (u) and correlation with sALT (healthy, n = 6; NASH, n = 10)(v). Cytotoxic activity results are representative of at least three independent experiments. Exact P (a, b, d, e, g–v) and n (n, o, t) values are presented in Source Data. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA with Sidak’s multiple comparison test (d, e), one-way ANOVA with Tukey’s (j, k, s, t) or with Dunnett’s (b, g, h) multiple comparison test and unpaired two-tailed t-test (a, i, l–n, p, r, u). In a, b, d, e, g–v, data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 9 Metabolic activation of CXCR6+ CD8 T cells triggers auto-aggression.
a, Phosphorylated proteins downstream of TCR signalling in IL-15-stimulated, acetate-exposed CXCR6+ CD8 T cells. b, Glycolysis in auto-aggressive CXCR6+ CD8 T cells (n = 3). c, Calcium influx in IL-15-stimulated CXCR6+ and CXCR6− CD8 T cells exposed to metabolites, cytokines or TLR ligands (n = 4). d, e, Extracellular ATP in primary mouse hepatocyte supernatant after 24 h treatment with palmitate (250 μM) or agonistic trimeric FasL, or coculture with auto-aggressive CD8 T cells (n = 3). f, g, Metabolite detection in liver tissue of mice fed a normal diet or CD-HFD by matrix-assisted laser desorption and ionization imaging. Scale bar, 500 μm (f). Exact minima, maxima, centres, bounds of box and whiskers and percentiles are presented in Source Data. h, P2RX7 expression in IL-15-stimulated CD44+CXCR6+ CD8 T cells. i, j, Calcium influx in CXCR6+ CD8 T cells after incubation with NAD (1 μM) (n = 5) or supernatants from d (n = 3) for 30 min. k, Representative cytotoxicity result of CD8 T cell auto-aggression in the presence of P2RX7 or PANX1 inhibition (n = 3). l, sALT at day 2 after transfer of auto-aggressive CD8 T cells and anti-P2RX7 nanobody (50 μg per mouse). Two independent experiments. m, Representative cytotoxicity result of antigen-specific, IL-15-stimulated and acetate-exposed CXCR6+ OT1 CD8 T cells against S8L-loaded hepatocytes. Exact P (a–e, i, j, l) and n (a, l) values are presented in Source Data. *P < 0.05, **P < 0.01, ****P < 0.0001. Two-way ANOVA with Tukey’s multiple comparison test (c, i, j), one-way ANOVA with Tukey’s (b, e, l) or with Dunnett’s (d) multiple comparison test and unpaired two-tailed t-test (a). In a–e, i, j, l data are mean ± s.e.m., error is reported as s.d.
Extended Data Fig. 10 Auto-aggressive human CD8 T cells.
a, Auto-aggression of allogeneic and autologous CD8 T cells against primary human hepatocytes after incubation with IL-15 (effector-to-target ratio of 3:1 (n = 3)). b, Auto-aggression by IL-15-treated human CD8 T cells exposed to acetate (n = 3). c, Acetate levels in livers from unaffected individuals or patients with NASH (n = 3). d, Auto-aggression of human CD8 T cells in presence of anti-TNF (10 μg ml−1) or anti-LFA1 (10 μg ml−1) (n = 3). e, f, Auto-aggression against MHC-I-deficient K562 cells by human CD8 T cells exposed to IL-15 (e), and at different effector-to-target ratios (n = 6). g, Calcium influx in IL-15-treated human CXCR6+CD69+ CD8 T cells measured with a calcium sensor dye before or after coculture with K562 cells and quantification (n = 6). h, i, Auto-aggressive killing of K562 cells by IL-15-treated CD8 T cells in presence of anti-FasL (10 μg ml−1), anti-NKG2D (10 μg ml−1) or EGTA (4 mM) at effector-to-target ratio of 10:1 (n = 6). j, Mean numbers of CD8 T cells adjacent to cleaved CASP3+ hepatocytes in mice fed a normal diet and NASH mice (n = 3). k, l, Surface FasL and TRAIL expression of mouse CXCR6+ CD8 T cells after 1 h exposure to ATP (200 μM). m, Caspase activity in primary mouse hepatocytes cocultured with auto-aggressive CD8 T cells and anti-FasL (10 μg ml−1) (n = 3). n, Mouse CD8 T cell auto-aggression in presence of inhibitors against apoptosis (zVAD-FMK, 10 μM) or necroptosis (Nec-1s, 30 μM) (n = 4). o, sALT at day 2 after adoptive transfer of IL-15-treated, acetate-exposed CD8 T cells and anti-FasL injection (100 μg per mouse). Two independent experiments. Exact P (a–o) and n (k, l, o) values are presented in Source Data. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Two-way ANOVA with Sidak’s (a) and Tukey’s (f, g) multiple comparison test, one-way ANOVA with Tukey’s multiple comparison test (b, d, e, h, i, m–o) and unpaired two-tailed t-test (c, j–l). In a–o, data are mean ± s.e.m., error is reported as s.d.
Supplementary information
Supplementary Data 1
Gating strategy of murine and human liver-infiltrating lymphocytes and patient characteristics of samples used in the study. a, Gating strategy of liver-infiltrating lymphocytes with exclusion of duplets and dead cells. b, Gating strategy of human liver-infiltrating lymphocytes with prior exclusion of duplets and live CD45+ cells as reference population for the frequency of CD69+CXCR6+ CD8 T cells in NAFLD/NASH patients. c, Patient characteristics including methods, cohort groups, gender, age, BMI, ALT, AST and NAS where available. d, Murine primer sequences for qPCR to quantify respective genes in the liver tissue of ND and CD-HFD mice.
Supplementary Information Table 1
Significant differentially expressed genes in liver CXCR6 positive compared to CXCR6 negative CD8 T cells from CD-HFD mice.
Supplementary Information Table 2
GSEA for differentially expressed genes in liver CXCR6 positive compared to CXCR6 negative CD8 T cells from CD-HFD mice.
Supplementary Information Table 3
Upstream transcription factors in CXCR6 positive compared to CXCR6 negative CD8 T cells from CD-HFD mice.
Supplementary Information Table 4
Differentially expressed genes in human CD8 T cells from steatotic livers.
Supplementary Information Table 5
Differentially expressed genes in human CD8 T cells from NASH livers.
Supplementary Information Table 6
Frequencies of human liver CD8 T cells from various clusters expressing PDCD1 or CXCR6 in steatotic and NASH livers.
Supplementary Information Table 7
Gene set enrichment analysis of differentially expressed genes of liver T cells from NASH patients for IL-15 dependent genes identified by Setoguchi et al 2016.
Supplementary Information Table 8
Gene set enrichment analysis of human liver T cells for T resident memory cell gene set identified by Zhao et al 2020.
Supplementary Information Table 9
Cluster analysis of CD3+ T cells from single cell RNA sequencing of CD3+ T cells from fat tissue of NASH patients.
Supplementary Information Table 10
Comparison of human liver and fat CD8 T cells from NASH patients analyzed by single cell RNA sequencing.
Supplementary Information Table 11
Commonly regulated genes in human RGS1+ and GNLY+FGFBP2+ CD8 T cells from NASH patients compared to differentially expressed genes in CXCR6 positive in comparison to CXCR6 negative CD8 T cells from NASH mice.
Supplementary Information Table 12
Gene set enrichment analysis of acetate- and butyrate dependent genes in CXCR6 positive in comparison to CXCR6 negative CD8 T cells from NASH mice.
Source data
Rights and permissions
About this article
Cite this article
Dudek, M., Pfister, D., Donakonda, S. et al. Auto-aggressive CXCR6+ CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449 (2021). https://doi.org/10.1038/s41586-021-03233-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03233-8
This article is cited by
-
Translational research on drug development and biomarker discovery for hepatocellular carcinoma
Journal of Biomedical Science (2024)
-
The immunopathogenesis of narcolepsy type 1
Nature Reviews Immunology (2024)
-
Targeting pathogenic CD8+ tissue-resident T cells with chimeric antigen receptor therapy in murine autoimmune cholangitis
Nature Communications (2024)
-
Child–Pugh grade deterioration stratified by the etiology after transcatheter arterial chemoembolization as initial treatment for hepatocellular carcinoma
Scientific Reports (2024)
-
Bystander activated CD8+ T cells mediate neuropathology during viral infection via antigen-independent cytotoxicity
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.