Abstract
The molluscs Lucinoma capensis, Lembulus bicuspidatus and Nassarius vinctus are highly abundant in Namibian oxygen minimum zone sediments. To understand which nutritional strategies allow them to reach such impressive abundances in this extreme habitat we investigated their trophic diversity, including a chemosymbiosis in L. capensis, focussing on nitrogen biochemical pathways of the symbionts. We combined results of bulk nitrogen and carbon (δ13C and δ15N) and of compound-specific isotope analyses of amino acid nitrogen (AAs—δ15NPhe and δ15NGlu), with 16S rRNA gene sequencing of L. capensis tissues and also with exploratory results of ammonium, nitrate and nitrite turnover. The trophic position (TP) of the bivalve L. capensis is placed between autotrophy and mixotrophy, consistent with its proposed symbiosis with sulfur-oxidizing Candidatus Thiodiazotropha sp. symbionts. The symbionts are here revealed to perform nitrate reduction and ammonium uptake, with clear indications of ammonium host-symbionts recycling, but surprisingly unable to fix nitrogen. The TP of the bivalve L. bicuspidatus is placed in between mixotrophy and herbivory. The TP of the gastropod N. vinctus reflected omnivory. Multiple lines of evidences in combination with current ecosystem knowledge point to sedimented diatoms as important components of L. bicuspidatus and N. vinctus’ diet, likely supplemented at times with chemoautotrophic bacteria. This study highlights the importance of benthic-pelagic coupling that fosters the dietary base for macrozoobenthos in the OMZ. It further unveils that, in contrast to all shallow water lucinid symbionts, deeper water lucinid symbionts rely on ammonium assimilation rather than dinitrogen fixation to obtain nitrogen for growth.
Similar content being viewed by others
Introduction
The Namibian shelf is one of the most productive upwelling areas in the world characterized by an enduring organic matter turnover, which leads to the formation of an oxygen minimum zone (OMZ) within the Benguela upwelling system (BUS). Shelf sediments shallower than 150 m are characterized by organic matter-rich diatom mud belts, where sulfur bacteria mats may form due to periodic formation of hydrogen sulfide (H2S)1,2,3. The combination of low oxygen availability and accumulations of toxic hydrogen sulfide in the sediment present extreme conditions for life2,3,4, and the central region of the OMZ supports low benthic density, biomass and highly variable dominant taxa (e.g. polychaetes, crustaceans, molluscs)5. However, the borders of the OMZ are home to a remarkably high biomass of mollusc species, including the bivalve Lembulus bicuspidatus and the gastropod Nassarius vinctus6. Furthermore, during a 2019 expedition (M157), we discovered an extraordinarily high biomass of the bivalve Lucinoma capensis (mean and maximum biomass of 50 g m−2 and 300 g m−2, respectively; Supplementary Table S1) in the biogenic mud neighbouring areas with the highest diffusive hydrogen sulfide fluxes at the sediment surface in the BUS2.
Members of the bivalve family Lucinidae have a global distribution and all species studied thus far form a nutritional symbiosis with environmentally acquired sulfur-oxidizing gammaproteobacteria, which may also play an important role in detoxifying hydrogen sulfide for their host7,8,9. Most shallow-water lucinids studied to date host symbionts from the genus Candidatus Thiodiazotropha10,11,12,13, with the exception of the mangrove lucinid Phacoides pectinatus which hosts a symbiont from the genus Sedimenticola14. In addition to the oxidation of reduced sulfur compounds, like hydrogen sulfide, and carbon fixation, all the various Ca. Thiodiazotropha symbiont species have the capacity to assimilate ammonium (NH4+) and to fix atmospheric nitrogen (N2), but vary in their ability to assimilate nitrate (NO3−) and urea, which seems to be a strategy for living in nitrogen-limited shallow-water environments (e. g. seagrass meadows and coral reefs)10,11,12. Despite growing interest in the chemosynthetic symbiosis of shallow-water lucinids, our understanding of the biology of the lucinids that inhabit the sediment of outer shelves and slopes remains very limited15,16. The deep water chemosynthesis-based ecosystems are often seeps and OMZs, which are environments characterized by distinct inorganic nitrogen cycling such as prominent ammonium efflux from the sediments due to reducing conditions17.
The molluscs L. bicuspidatus and N. vinctus also dominate the Namibian shelf sediments6. The bivalve L. bicuspidatus (Nuculanidae) is an infaunal deposit feeder with limited mobility that inhabits organic-rich sediments6,18. An unexpectedly high density of L. bicuspidatus occurs northwards of the L. capensis populations at a mean abundance and biomass of 515 individuals and 257 g m−2, respectively (Supplementary Table S1)6. N. vinctus is a highly mobile gastropod known for scavenging on dead animals and tolerance of a wide range of ambient oxygen levels3,19. N. vinctus co-occurs with L. capensis and L. bicuspidatus but lives on the sediment surface where it reaches high abundances (328 individuals m−2) and makes up for less of the total mollusc biomass (37 g m−2) (Supplementary Table S1). Therefore, despite its inhospitality, the OMZ of the BUS supports a surprisingly high biomass of benthic animals, yet little is known about their trophic ecology or the nutrient sources that allow them to achieve such high biomass under the harsh conditions in the OMZ20.
Stable isotopic analysis (SIA) has been widely used to study the trophic interactions of marine benthic communities19,21,22,23, and is a powerful tool for detecting nutrient sources and tracing nutrient exchanges in chemoautotrophic associations24,25,26. For example, the bulk δ13C values of chemosymbiotic lucinids (− 37‰ to − 23‰)27,28,29,30,31 are approximately within the bulk δ13C range expected when a large proportion of their carbon is derived from autotrophy with the CO2-fixing enzyme Rubisco I (− 35‰ to − 27‰)24. In contrast, non-symbiotic benthic organisms typically have δ13C values between − 20 and − 16‰29,32. Specifically, a non-symbiotic generalist deposit feeder, for instance, is expected to have δ13C values that resemble the signature of the local sedimentary organic matter or free-living bacteria32. Interestingly, the δ13C values of non-symbiotic benthic organisms appear to reflect signatures resulting from activities of the CO2-fixing enzymes PEP carboxylase and PEP carboxykinase in marine phytoplankton33, which may indicate that marine phytoplankton is a dietary base for non-symbiotic benthic food webs.
Bulk δ15N values vary from − 13 to 5‰ in chemosymbiotic organisms, and typically from 5‰ to 15‰ in non-symbiotic benthic organisms30,32,34,35,36,37,38,39. Bulk δ13C values and bulk δ15N values are often combined in SIA, however bulk δ15N values are affected by higher and more variable trophic fractionation between diet and consumers than bulk δ13C values (∆δ13consumer-diet of 1.0‰). Bulk δ15N values are enriched by 2.3–3.4‰ in consumers compared to their diet, and more specifically by 2.5‰ ± 2.5‰ in herbivores and by 3.2‰ ± 0.4‰ in carnivores40,41. Therefore, one might expect the bulk δ15N of a deposit feeder inhabiting the Namibian shelf to reflect the δ15N signature of the local surface sediment nitrogen of 4.6–10.0‰42,43, albeit trophically enriched by 2.3–3.4‰.
However, accurate interpretation of bulk stable isotopes values requires prior characterization of the dietary base line, which is not always available44. Compound-specific isotope analyses of amino acid nitrogen (CSIA-AA) is a refining approach allowing the more precise assignment of trophic positions (TP) without prior base line characterization45,46,47. This is achieved by comparing the δ15N value of the so-called “trophic AA” glutamic acid (Glu), which is enriched by 6.4‰ ± 2.5‰ per trophic transfer, with the δ15N value of the so-called “source AA” phenylalanine (Phe), which reflects the isotopic composition of the dietary base line because its δ15N value is marginally fractionated during trophic transfer by only − 0.1‰ ± 1.6‰45,47,48. Because of the conservative nature of the δ15N value of phenylalanine in the food web, it is possible to further explore the incorporation of pelagic diatoms (sedimented over the sea floor) by members of the benthic food web by comparing the δ15NPhe values of benthic consumers with the δ15NPhe values of herbivorous mesozooplankton from the Namibian shelf, which has a δ15NPhe value of 5.3‰49 (Supplementary Table S2).
Besides trophic interactions studies, nitrogen stable isotopes appear to reflect signatures from the dominant inorganic nitrogen source assimilated by the autotrophic base of a food web, when considering the isotopic fractionation of the specific nitrogen assimilation process. That can be done by comparing the bulk δ15N of an autotrophic base of a food web, with the δ15N values of the local dominant inorganic nitrogen sources available, which are namely dinitrogen, nitrate and ammonium in both bottom and surface water off Namibia. For dinitrogen, we consider the δ15N to be 0.6‰ as for dinitrogen dissolved in seawater, and the maximum isotopic fractionation during dinitrogen fixation to be 2.5‰50. Thus, if autotrophic organisms would primarily grow on dinitrogen fixation, their bulk δ15N in particulate organic nitrogen (PON) would have a minimum value of − 2.1‰ (Supplementary Table S2).
The theoretical values of δ15N of nitrate assimilating PON in the Namibian shelf must consider two sources of extremes for δ15N nitrate, the upwelling water and the sediment-overlying water. The source water for upwelling in the northern BUS originates from South Atlantic Central Water (SACW), that feeds into an undercurrent off the shelf break, whose nitrate δ15N values vary between 5.7 and 6.7‰ near the central shelf edge (~ 170 m)17. From there, the upwelling water is transported towards the Namibian shelf reaching surface waters and providing comparatively isotopically lighter nitrate for pelagic phytoplankton growth, namely of diatoms. On the other hand, the suboxic or anoxic sediment-overlying water is a second isotopically heavier local nitrate source with δ15N values of 5.9–14.3‰, that are thought to be mainly due to isotope fractionation of nitrate during denitrification spatially overlapping with low oxygenated conditions17,51. The isotopic contrast between pelagic and benthic nitrate sources may be reflected in enriched δ15N values of PON in nitrate assimilating benthic chemoautotrophs and its consumers compared to more depleted δ15N at the dietary base of the pelagic food web (Supplementary Table S2). Thus, considering the isotopic fractionation to range between 4.0 and 10.0‰50 for nitrate assimilation by photoautotrophs, phytoplankton growing on nitrate assimilation from upwelled waters would have bulk δ15N for PON ranging between − 4.3 to 2.7‰ in the Namibian shelf (Supplementary Table S2). To the best of our knowledge, the fractionation factor associated with growth on nitrate in chemoautotrophic bacteria is unknown. Thus a first approximation may be to assume a similar fractionation factor as in nitrate assimilation by other prokaryotes (0.4‰ to 5.0‰)52. In that case, benthic chemoautotrophic bacteria growing on a local, shelf-specific nitrate source would have bulk δ15N for PON ranging between 0.9 and 13.9‰ at the central shelf (Supplementary Table S2).
Ammonium is abundant in the sediments of the Namibian shelf, and thus a promising inorganic nitrogen source for the benthic ecosystem. To the best of our knowledge, the δ15N of ammonium has not been measured for our study area. However, assuming that the δ15N of ammonium largely reflects the δ15N of particulate organic nitrogen settling to the seafloor, and that δ15N fractionation during ammonification of PON in organic-rich marine sediments is negligible53,54,55, we can ascribe the δ15N values of ammonium as the same values of sediment PON (Supplementary Table S2). Finally, knowing that the assimilation of ammonium can be associated with isotope fractionation varying from 5 to 20‰56, it is possible to estimate the δ15N of ammonium assimilating PON.
In this way, we will deduce which inorganic nitrogen source is being assimilated by the L. capensis symbionts by excluding the processes that cannot explain the bulk δ15N values in the gill tissue from L. capensis. Also, the more refined δ15NPhe values will assist us on identifying dietary sources from, for instance, pelagic realms, which may strongly support benthic environment.
Our general aim was to investigate the trophic ecology of three dominant mollusc species in the Namibian shelf. We analysed bulk δ13C and bulk δ15N values as well as amino acid nitrogen specific δ15NPhe and δ15NGlu values of their gill (in case of L. capensis) and body tissues. These values are then compared to the estimated (in case of ammonium and diatoms’ PON) and reference (in case of dinitrogen, nitrate, sediment PON) δ15N values of the dominant organic and inorganic nitrogen pools in the OMZ including diatoms’ PON, sediment PON, dinitrogen, nitrate, and ammonium. Doing so allowed us to infer the mean trophic position (TP) of all species, the dominant inorganic nitrogen source actively used by the symbionts in case of L. capensis, and the main food sources of all three molluscs on the Namibian shelf17,19,21,22,23,24,26,43. We hypothesized that (i) the values of bulk δ13C, bulk δ15N, δ15NPhe, δ15NGlu and TP of L. capensis will be the lowest among the three species and consistent with an autotrophic lifestyle through their chemoautotrophic symbionts that grow mainly on ammonium, (ii) L. bicuspidatus will have intermediate values consistent with omnivory and the consumption of surface sediment PON, and (iii) N. vinctus would have the highest values, indicating carnivory and consistent with the scavenging of other (namely dead) animals. We combined our isotope based approach with ammonium, nitrate, nitrite and oxygen flux measurements, and metagenomic sequencing of the L. capensis gills to robustly unravel the nitrogen biochemical pathways and potential nitrogen sources (dinitrogen, nitrate or ammonium) that sustain the L. capensis symbiosis in the OMZ off Namibia.
Methods
Sampling
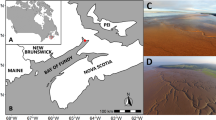
Lucinoma capensis, Lembulus bicuspidatus and Nassarius vinctus were collected on board the RV Meteor during the EVAR Expedition M157 (18.08.2019–14.09.2019), by dredging at different stations in the Northern Benguela Upwelling System (Fig. 1, Table 1). Species were found at different stations (Stns.) possibly due to large environmental differences among stations. L. capensis individuals were collected in the middle and southern part of the investigation area at Stns. 12 and 48 (central and southern Namibian shelf) (Table 1). L. bicuspidatus were collected in the northern part at Stns. 24 and 30 (northern Namibian shelf). N. vinctus individuals were collected in the middle part of the investigation area at Stns. 14 and 12 (central Namibian shelf), although they can be found anywhere along the shelf. Additionally, sediment samples were collected at Stns. 14 and 48 (central and southern Namibian shelf) for bulk carbon and nitrogen stable isotope analysis. The oxygen concentration at the benthic boundary layer was measured by CTD-profiling with an attached oxygen sensor. Sediment cores were retrieved at Stns. 12, 24, 30 and 48, and ammonium and nitrate concentrations in the water overlying the sediment at these sites were measured on an auto-analyzer (QuAAtro, Seal analytical) using standard colorimetric methods by Grasshoff et al.57. We used sediment bulk δ15N PON values from Stn. GB1 (GeoB 1001-l-RV Meteor 6/6 1988) from Holmes et al.43. We obtained δ15N values of nitrate for the sediment-overlying water in the central Namibian shelf by using the data of Stns. 198 and 252 from Nagel et al.51, which are the extreme values measured (Supplementary Table S2).
Locations of stations sampled during the M157 cruise (24, 30, 12, 14, 48). We additionally plotted the station GB1 (GeoB 1001-l-RV Meteor 6/6 1988) from Holmes et al.43, from which we used sediment bulk δ15N PON data, Stns. 252 and 198 from Nagel et al.51, from which we used δ15N of nitrate data. The letters next to the station code refer to which samples were collected or data were used in this study: s—sediment; b—L. bicuspidatus; c—L. capensis; v—N. vinctus; N—δ15N of nitrate; Figure created using ESRI ArcGIS 10.8.1. Final layout of the figure was done in Inkscape 1.1.2, www.inkscape.org.
Bulk stable carbon and nitrogen isotope analyses
We performed bulk stable carbon and nitrogen isotope analyses with tissues of L. capensis, L. bicuspidatus, N. vinctus and surface sediment samples, aiming to identify the main food sources of the molluscs and also the inorganic nitrogen source assimilated by symbionts. The δ15N values from the sediment surface will also be used for estimating δ15N of ammonium, by assuming that δ15N fractionation during ammonification of PON in organic-rich marine sediments is negligible53,54,55. A total of three individuals of L. capensis from Stn. 48, 13 individuals of L. bicuspidatus from Stn. 24, and six individuals of N. vinctus from Stn. 12 were used for bulk stable isotope analysis. Specimens were collected, freshly and entirely snap frozen and stored at − 80 °C until freeze dried. L. capensis gills were separated from the non-symbiotic tissue. Gut content was not purged, yet we assume that the volume of the gut was negligible compared to the volume of the tissues, and gut content may not have largely affected animals’ isotopic values. The dry frozen tissues were carefully removed from the shells and ground to a homogeneous fine powder. Approximately 0.5 mg of each sample was weighted into silver weighing boats. The samples were either acidified (HCL) or non-acidified prior to bulk δ13C (in the following δ13CBulk) and bulk δ15N (in the following δ15NBulk) analyses (Supplementary Table S3). The non-acidified subsamples were additionally used for amino acid nitrogen specific isotope analysis.
The bulk analyses were performed at the EA-IRMS Laboratory of the Leibniz Institute for the Baltic Sea Research, Germany. Carbon and nitrogen stable isotope ratios were measured using an Elemental Analyzer (EA, Thermo Flash EA 1112), a continuous flow isotope ratio mass spectrometer (IRMS, Thermo Finnigan DeltaPlus) via an open split interface (Thermo Finnigan Conflo IV). The nitrogen (N2) and carbon dioxide (CO2) reference gases for stable δ15N and δ13C isotope analyses were calibrated against the reference materials from the International Atomic Energy Agency (IAEA): IAEA-N3 (KNO3) and IAEA-CH-6 (sucrose). The control of isotopic composition was done by a peptone in-house standard (Merck) after every sixth sample. The peptone standard indicated an analytical error associated with the isotope measurements of less than 0.2‰ for both isotopes.
Amino acid nitrogen specific isotope analysis
We performed amino acid nitrogen specific isotope analysis with tissues of L. capensis, L. bicuspidatus and N. vinctus to better determine their trophic position and their main food source. The amino acid compound specific isotope analysis45,46 was performed with 10 mg subsamples of the powdered gill and non-symbiotic tissue of three L. capensis individuals from Stn. 48, three whole N. vinctus individuals from Stn. 12, three whole L. bicuspidatus individuals from Stn. 24. The δ15N values of triflouroacetyl/isopropyl ester (TFA) AA derivatives58,59 were measured on a Thermo GC Isolink CN with Trace 1310 (GC), coupled via a ConFlo IV combustion interface (C) to a MAT 253 (IRMS, all Thermo Fisher Scientific GmbH, Dreieich, Germany) in Warnemünde. The precision of our GC-C-IRMS measurements varied among individual AAs and sample type but the standard deviation of 2–5 analyses typically was ≤ 1.0‰. Here we show the δ15N values for glutamic acid (in the following δ15NGlu) and phenylalanine (in the following δ15NPhe). Details of the analysis can be found in Loick-Wilde et al.60.
Biogeochemical measurements
The trophic positions (TP) were calculated according to the following equation61:
with β = 3.4 and ΔGlu-Phe = 7.662. A TP value of 1.0 indicates autotrophy, 2.0 indicates herbivory and 3.0 indicates the first level of carnivory46,48. TP values within the range of 1.0–2.0 indicate mixotrophy, while values within the range of 2.0–3.0 indicate omnivory63,64,65,66. Depending on environmental conditions, the TP values of mixotrophic organisms may alternate between phases that are mainly “autotrophic”, with TP values of approximately 1.3, or mainly “herbivory”, with TP values of approximately 1.967. Standard error (SE) in TP estimations, computed by propagation of analytical error in the individual AA determinations, did not exceed 0.2 TP.
On board rate measurements of oxygen, nitrate, nitrite and ammonium
To support our assumptions regarding inorganic nitrogen assimilation by L. capensis symbionts, we have considered the results of exploratory incubations in which oxygen, nitrate, nitrite and ammonium were measured over time in the presence of each mollusc. Freshly sampled specimens were incubated on board the FS Meteor for metabolic rate measurements. Five airtight vials (20 ml) were filled with unfiltered sea-water from the benthic boundary layer at Stn. 12; seawater was collected with 10 l free-flow bottles (Hydrobios, Germany) attached to a rosette water sampler. One N. vinctus (13–14 mm in size) from Stn. 14 was placed in each of 3 vials; incubations were performed under approximately in situ starting conditions (10 °C). Oxygen was monitored until the first vial became anoxic (after 52 h). Another five airtight vials were filled with unfiltered water taken from cores collected at Stn. 12 by retrieving a multicore, and L. capensis specimens (9–12 mm in size) from Stn. 12 were incubated one individual per vial (n = 3) under approximately in situ environmental conditions (10.4 °C). Oxygen was monitored until the first vial became anoxic (after 94 h). Another four airtight vials were filled with non-filtered water removed from cores collected at Stn. 30 by retrieving a multicore. Two of the vials had one individual of L. bicuspidatus (size between 13 and 15.5 mm) from Stn. 30; incubations were performed under approximately in situ environmental conditions (14 °C). Oxygen was monitored until the first vial became anoxic (after 51 h). For all incubation experiments described above, one extra vial was used as blank, and another for temperature measurements. Nitrate, nitrite and ammonium concentrations were measured at the end of the incubations.
Nitrite, nitrate and ammonium concentrations were measured on an auto-analyzer (QuAAtro, Seal analytical) using standard colorimetric methods by Grasshoff et al.57. The nutrient samples were previously stored at 4 °C for no longer than two days. Oxygen concentrations were measured through the Optical Oxygen Meter FireStingO2 (4 channels) (PyroScience GmbH, Germany) connected with optical fibers (SPFIB) and one temperature sensor (SPFIB), using contactless oxygen sensors spots (OXSP5) attached with silicon (SPBLUE). The oxygen and the nutrients net consumption/production were calculated by subtracting the blank values from the non-blank values, divided by incubation period and multiplied by the water volume (V) using the following equation:
The final value was divided by the Ash Free Dry Weight (AFDW) of the organism, in order to standardize the rates. We calculated wet weight by using a model of size x wet weight for each species. We further converted the wet weight values to AFDW by using the IOW-Benthic Ecology Laboratory internal conversion factors of Nassarius incrassatus (0.081), Lucinoma borealis (0.091) and Ennucula tenuis (0.084) for N. vinctus, L. capensis and L. bicuspidatus, respectively.
Nucleic acid extraction and sequencing
Gill tissues were rapidly dissected from frozen L. capensis specimens. Genomic DNA (gDNA) and total RNA was extracted from approximately 300 mg tissue using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s procedures with the following modifications: The tissue was cut into pieces, vortexed with silanised glass beads (2 mm and 3 mm), 1 ml solution of 99.99% β-mercaptoethanol and 1% of RLT Plus buffer for 15 min. The resulting solution was then distributed into two tubes, diluted to 50% Beta merkaptor and 50% RLT Plus buffer. The extracted DNA (86.92 ng/µl ± 14.91) and RNA (182.90 ng/µl ± 89.20) were quantified with a NanoDrop ND‐1000 spectrophotometer (NanoDrop Technologies, Wilmington, Delaware, U.S.A.). cDNA was synthesized from the RNA extracts using Multiscribe reverse transcriptase, according to manufacturer’s instructions (Thermo Fisher Scientific # 4311235).
Metagenomic and amplicon sequencing were carried out by the Joint Microbiome Facility of the Medical University of Vienna and the University of Vienna under project IDs JMF-2104-13 and JMF-2104-02, respectively. The V3–V4 regions of the 16S rRNA genes were amplified from both the DNA and cDNA of RNA extracts using the 341F and 785R primers. Library preparation, sequencing, and analysis of the amplicon sequence variants (ASVs) were carried out as described by Pjevac et al.68. DNA libraries for metagenome sequencing were prepared using the Illumina compatible NEBNext Ultra II FS DNA Library Prep Kit. The libraries were sequenced on an Illumina NovaSeq 6000 using paired-end settings for read-lengths of 100 bp to generate a minimum of 1,000,000 reads.
Quality filtering, genome assembly, and bacterial genome binning were carried out using the scripts and workflow previously described by Osvatic et al.13. Only MAGs with over 90% completion (CheckM gammaproteobacterial gene set) and lower than 5% contamination were kept in subsequent downstream analysis. The taxonomy of all MAGs was assessed using GTDB version 0.3.3 (X). MAGs assigned to the genus Ca. Thiodiazotropha were submitted to the RAST (Rapid Annotation using Subsystem Technology; https://rast.nmpdr.org/) web server for functional annotation using the default RASTtk pipeline69.
Data and analysis deposition
The data (raw reads and MAGs) have been deposited with links to BioProject accession numbers PRJNA765502 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). The BioSample accession numbers for the MAGs are SAMN21561986-SAMN21561988 and SAMN21572993-SAMN21572995 are for the corresponding raw read sets.
Results
Bulk carbon and nitrogen isotopic signatures of L. capensis, L. bicuspidatus, N. vinctus and the surrounding sediments
The δ13CBulk values of the body and gill tissues of the three mollusc species and sediment samples spanned a wide range from − 30.8 to − 16.3‰, with the very low values for the species L. capensis and similar high values for the species L. bicuspidatus and N. vinctus (Fig. 2). Specifically, the average δ13CBulk value of the L. capensis tissues was the lowest of the three molluscs with − 29.8‰ ± 0.9‰ (n = 3) in the symbiont-housing gills and − 28.8‰ ± 0.8‰ (n = 3) in the non-symbiotic tissue (Fig. 2). In contrast, the average δ13CBulk values for tissues of the bivalve L. bicuspidatus and the gastropod N. vinctus were much higher at − 16.7‰ ± 0.4‰ (n = 13) and − 17.5‰ ± 0.1‰ (n = 6), respectively. The δ13CBulk values of the surface sediments had intermediate values but were clearly more depleted (> 1.0‰ difference) than those of the non-symbiotic mollusc species. Specifically, δ13CBulk values of the sediments were − 20.3‰, − 20.4‰, and − 21.1‰ at the central (Stn. 14), southernmost (Stn. 48), and northernmost stations (Stn. GeoB 1001-l)43, respectively.
Bulk δ15N (‰ vs. N2) and bulk δ13C (‰ vs. PDB) signatures of L. bicuspidatus (n = 13), N. vinctus (n = 6), L. capensis (gills and non-gills tissue) (n = 3), and sediments from Stations 14, 48, and GeoB1001-l43 (n = 1). Vertical lines show the δ15N range of nitrate in the sediment-overlying water at the central Namibian shelf (parallel line) (Stations 198 and 252)51 as well as in the upwelled SACW off the Namibian shelf (dotted parallel line)17. In addition, vertical help lines (dashed parallel line) show the estimated δ15N range for ammonium calculated from PON of St. 14, Stn. 48 and Geob 1001-l (Supplementary Table S2). The light blue square (blue filled square) and the light red square (red filled square) have δ15NBulk values of acidified samples which were corrected by subtracting 0.3‰, the difference between acidified and non acidified samples measured in this mollusc group that were both acidified and non acidified (Supplementary Table S3).
δ15NBulk average values of the body and gill tissues of the three mollusc species and sediment samples also covered a wide range from − 5.7 to 10.8‰, with lowest values for the species L. capensis, the highest values for the species N. vinctus, and intermediate values for L. bicuspidatus and sediments (Fig. 2). Specifically, the average δ15NBulk values of the L. capensis tissues were − 4.9‰ ± 0.7‰ (n = 3) in the gills and − 2.5‰ ± 0.6‰ (n = 3) in the non-symbiotic tissues. Average δ15NBulk values for the bivalve L. bicuspidatus and the gastropod N. vinctus were much higher than those of L. capensis, at 5.0‰ ± 0.3‰ (n = 13) and 10.4‰ ± 0.4‰ (n = 6), respectively. δ15NBulk value of sediment at the central station (Stn. 14) was 5.6‰ and thus slightly higher than the sediment of the southernmost (Stn. 48) and northernmost stations (Stn. GeoB 1001-l), which were 4.3‰ and 4.6‰43, respectively. Ammonium was considered to have the same δ15N values as sediment PON, thus 5.6‰ at the central station, 4.3‰ at the southernmost station and 4.6‰ at the northernmost stations (Supplementary Table S2).
The reference values from literature for nitrate in the sediment-overlying water at the central Stns. 198 and 252 range from 5.9 to 14.3‰51, respectively. The reference values from literature for nitrate in the upwelled SACW vary in a narrow range from 5.7 to 6.7‰17.
In summary, the > 1‰ differences in δ13CBulk values between sediment and all molluscs indicate no strong dietary connection between the two, e.g. if at all, only selective parts of the sediment not reflected in the bulk δ13C values may be incorporated. The higher δ15NBulk values in N. vinctus compared to L. bicuspidatus could be due to N. vinctus occupying a higher trophic position or due to an isotopically heavier dissolved inorganic nitrogen source sustaining the food webs in the oxygen depleted middle part of the study area, where N. vinctus was sampled. This will be resolved by the δ15NPhe and δ15NGlu data below, although the very similar δ13CBulk values between both molluscs already suggest an isotopically similar dietary source for L. bicuspidatus and N. vinctus.
Nitrogen sources and food web structure of L. capensis, L. bicuspidatus and N. vinctus
The body tissues of all three mollusc species, and the gills of L. capensis, were analyzed with CSIA-AA to determine their nitrogen sources (δ15NPhe) and trophic positions in the food web (TP). The δ15NPhe values of gill and body tissue samples revealed large differences among the species that were consistent with the δ15NBulk measurement patterns (Fig. 3). The δ15NPhe values in L. capensis from both gill and body tissues were clearly lower than the values in L. bicuspidatus and N. vinctus and ranged from − 7.7 to − 4.0‰ with a mean value of − 6.0‰ ± 1.8‰ (n = 3) and − 4.8‰ ± 1.0‰ (n = 3) for gill and body tissues, respectively.
δ15NGlu and δ15NPhe measured for L. capensis, L. bicuspidatus and N. vinctus. Trophic isoclines with a slope of 1.0 and a y-intercept interval of 7.6 represent different trophic positions (TPs = 1.0, 1.5, 2.0, 2.5, and 3.0)63,64,65,66. The “mainly autotrophy” and “mainly herbivory” TP lines were estimated from the range of nutritional states experienced by the mixotrophic kleptoplastic gastropod Plakobranchus ocellatus Maeda et al.67. The δ15NPhe of PON of sedimented diatoms for the Namibian shelf (dashed line) were back calculated from mesozooplankton δ15NPhe data of Steinkopf49 (Supplementary Table S2).
The δ15NPhe values in L. bicuspidatus and N. vinctus had similar mean values of 4.7‰ ± 2.6‰ (n = 3) and 6.4‰ ± 0.2‰ (n = 3), which supports the δ13CBulk data indicating that both species have isotopically similar diets. Further, it shows that the differences in δ15NBulk values between both species (Fig. 2) cannot be explained by a change in the δ15N of the dietary baseline. Moreover, those values are close to the PON δ15NPhe value estimated for sedimented diatoms consumers (5.3‰)49 (Supplementary Table S2) in the shelf.
The estimated TPs of the L. capensis gill and body tissue samples using the TPGlu/Phe approach ranged from 1.1 to 1.7 (Fig. 3), thus mostly reflecting material of autotrophic and mixotrophic origin64,65,70. In contrast, the TP of L. bicuspidatus ranged from 1.4 to 1.9 (Fig. 3) reflecting mainly herbivorous and some mixotrophic feeding behaviors. Interestingly, no clear signal of predominantly carnivorous feeding behavior (TP ≥ 3.0) was found in the putative scavenger N. vinctus, whose TP values ranged from 2.4 to 2.7 (Fig. 3) reflecting mainly omnivorous feeding behaviors. Further, the TP comparison between L. bicuspidatus and N. vinctus shows that the differences in δ15NBulk values between both species (Fig. 2) can be explained by the clearly higher TP of N. vinctus (Fig. 3).
The Lucinoma capensis gills harbour bacterial symbionts from the genus Candidatus Thiodiazotropha
A single ASV from the genus Candidatus Thiodiazotropha dominated the L. capensis gill microbiome (ASV ID: ASV_6cc_309) (Fig. 4). This ASV made up between 93 and 98% of all amplicons from the DNA fraction and 98–100% of amplicons from the cDNA fractions. ASVs belonging to Spirochaetes, which have also previously been reported in lucinid gill microbiomes, were detected in the amplicon libraries from DNA, with relative abundance between 0.1 and 3%. In cDNA spirochaete ASVs made up less than 0.1% of the amplicons. Imaging methods such as FISH would be needed to demonstrate conclusively that the Ca. Thiodazotropha ASV detected in these animals is an intracellular symbiont like all other Ca. Thiodiazotropha so far investigated. However, it is likely that this highly abundant ASV, related to other lucinid symbionts, is also a gill symbiont of L. capensis. To further investigate the functional capabilities of this potential Ca. Thiodiazotropha symbiont, we sequenced, assembled, and binned the gill metagenomes of three L. capensis individuals. We assembled three MAGs (metagenome assembled genomes) that were assigned to the Gammaproteobacteria genus Candidatus Thiodiazotropha. The assembled MAGs ranged in size from 4.40 to 4.56 megabases pairs and together represented a single symbiont species based on an ANI (average nucleotide identity) threshold of 95%, and they share a 78% ANI with Ca. Thiodiazotropha endolucinida. All three MAGs were considered high quality with > 99% completeness and < 5% contamination.
Multiple pathways for oxidizing reduced sulfur compounds were encoded in the MAGs of the L. capensis symbionts (Table 2), all of which are similar to those present in previously described lucinid symbionts10,12,13,14. Energy gained from sulfur oxidation is used to fix inorganic carbon through the Calvin–Benson–Bassham (CBB) cycle, and the L. capensis symbiont MAGs encoded genes for two distinct types (forms I and II) of Ribulose-1,5-bisphosphate carboxylase/oxygenase. The L. capensis symbiont MAGs encoded genes required for heterotrophic growth, ammonium assimilation and the ability to use nitrate (narGHIJ), and nitrite (nirBD) as alternative electron acceptors like many of the previously published genomes. The genus name Thiodiazotropha was proposed in reference to the ability of shallow-water lucinid symbionts to fix inorganic nitrogen from the atmosphere11,12. However, we did not detect any genes involved in nitrogen fixation or genes involved in converting urea to ammonium; both nitrogen fixation and urea degradation are predicted functional capabilities encoded in many lucinid symbiont genomes. The absence of nitrogen fixation genes is surprising, as this pathway is present in virtually all so-far sequenced lucinid symbionts.
Nitrogen compound flux measurements
Nitrate net consumption (Fig. 5a) was highest in incubations with L. capensis (− 0.16 ± 0.002 µmol h−1 AFDW gram−1, n = 3) compared to N. vinctus (− 0.05 ± 0.01 µmol h−1 AFDW gram−1, n = 3), while L. bicuspidatus did not consume nitrate (0.005/0.008 µmol h−1 AFDW gram−1, n = 2). Ammonium net production (Fig. 5b), was lowest in incubations with L. capensis (0.14 ± 0.1 µmol h−1 AFDW gram−1, n = 3) compared to N. vinctus (1.14 ± 0.10 µmol h−1 AFDW gram−1, n = 3), and L. bicuspidatus (0.91/1.03 µmol h−1 AFDW gram−1, n = 2). Oxygen net consumption (Fig. 5c) rates were similar between L. capensis and L. bicuspidatus incubations with an average rate of − 0.03 (sd ± 0.007, n = 3) ml h−1 AFDW gram−1 for the former and values ranging from − 0.035 to 0.033 ml h−1 AFDW gram−1 for the latter, while N. vinctus consumed the most oxygen at an average rate of − 0.11 (sd ± 0.01, n = 3) ml h−1 AFDW gram−1. Finally, nitrite net production (Fig. 5d) was the highest in L. capensis incubations (0.14 ± 0.14 µmol h−1 AFDW gram−1, n = 3), compared to N. vinctus (0.03 ± 0.01 µmol h−1 AFDW gram−1, n = 3), and L. bicuspidatus (0.008/0.19 µmol h−1 AFDW gram−1, n = 2).
Discussion
Chemoautotrophic bacteria and/or freshly sedimented diatoms contribute to the diets of the most abundant molluscs in the BUS
The OMZ within the BUS off Namibia is characterised by periods of low oxygen availability and high levels of toxic hydrogen sulfide. Nevertheless it supports a remarkably high biomass of three benthic mollusc species: a putatively chemosymbiotic bivalve L. capensis, a putatively detritivorous bivalve L. bicuspidatus, and a putatively scavenger gastropod N. vinctus. Through the combination of stable isotope analyses with nitrogen compound flux measurements and metagenomic sequencing, we identified their main food sources, determine their trophic positions, and gained insights into the nitrogen metabolic pathways of L. capensis symbionts.
The range of δ13CBulk values of L. capensis (− 30.8‰ to − 27.8‰) fall within the range of δ13CBulk values (− 37‰ to − 23‰) reported for various lucinid species27,28,29,30,31, and are most comparable to the δ13CBulk measurements in the lucinid Lucinoma kazani (δ13CBulk of − 30.5‰ in the gills and − 28.2‰ in non-symbiotic tissues), which inhabits a cold seep environment in the Eastern Mediterranean31. As the difference in bulk δ13C between host and symbionts is normally minimal, because carbon is enriched by only 0.4–1.0‰ between each trophic level24,32, these depleted δ13CBulk values indicate lucinid biomass is made up of carbon originally derived from symbiont CO2-fixation through a RuBisCO enzyme71,72. The L. capensis gill symbionts belong to the genus Ca. Thiodiazotropha, the members of which have been described as sulfur-oxidizing chemoautotrophic symbionts of shallow-water lucinids. All three L. capensis symbiont MAGs encode genes for both forms I and II RuBisCO enzymes indicating the potential to convert inorganic carbon to organic carbon (Table 2). Assuming that the δ13C of DIC near bottom of Stn. 12 and 48 (measured during the M103 cruise) are − 0.27‰ and − 0.37‰ at 11.9 °C and 11.7 °C, respectively73, thus the theoretical values of δ13C of CO2 should be − 10.8‰ and − 11.0‰74,75 at L. capensis’ stations. Given the bulk δ13C values of L. capensis gills (− 29.0‰ to − 30.8‰) and that the isotopic values of bulk POC of CO2 fixing organisms in nature are typically 10–20‰ heavier than theoretical values71, the discrimination against CO2 of the symbionts’ Rubisco would be between 28.0 and 40.0‰. Taking into account that Rubisco form I has been described to discriminate with an ε of 22–29‰ and Rubisco II with an ε of 17.8–23‰71, Rubisco form I is probably more strongly influencing the δ13C POC of the symbionts.
The ability of the L. capensis gill symbionts to oxidize reduced sulfur compounds and fix inorganic carbon, along with the low δ13CBulk and δ15NBulk values of the L. capensis tissues, suggest these bivalves rely on the chemosynthetic metabolism of their bacterial symbionts for nutrition7,8,9,24,29,31. These lines of evidence are strongly supported by the δ15NGlu and δ15NPhe data, which place L. capensis tissues at a trophic position that lies between autotrophy and mixotrophy (Fig. 3). The average TPs of L. capensis gills and non-symbiotic tissues (1.3 ± 0.3 and 1.4 ± 0.3, respectively) were similar to that of the kleptoplastic gastropod Plakobranchus ocellatus (1.3 ± 0.1) when laboratory-induced starvation of the normally algivorous animals led to an increased reliance on autotrophic nutrition from sequestered chloroplasts67. Mixotrophy could be an important nutritional strategy for lucinids inhabiting sediments of a highly productive Oxygen Minimum Zone, as was described in another OMZ lucinid L. aequizonata76, because feeding on particulate organic matter may allow them to survive periods of low hydrogen sulphide conditions76. Further studies are required to investigate the origin and importance of the particulate matter in the L. capensis diet.
The δ13CBulk and δ15NBulk values of the putatively non-symbiotic molluscs L. bicuspidatus and N. vinctus were much higher than that of the chemosymbiotic lucinid L. capensis (Fig. 2). Yet, δ13CBulk values alone are insufficient evidence to conclude that these two benthic molluscs are non-symbiotic, or do not consume chemoautotrophic bacteria, as the range of δ13CBulk values for non-symbiotic benthic organisms (− 20.0‰ to − 16.0‰)29,32 overlaps with the wide range of δ13CBulk values (− 35.0‰ to − 9.0‰) that have been reported for chemoautotrophic bacteria24. This is further confounded by δ15NBulk data, which place N. vinctus δ15NBulk values within the range of non-symbiotic benthic organisms (5‰ to 15‰), and L. bicuspidatus at the threshold of separating chemosymbiotic (− 13‰ to 5‰) and non-chemosymbiotic benthic organisms30,32,34,35,36,37,38,39. This scenario supports the statement that N. vinctus is not a symbiotic mollusc, however raises the question whether L. bicuspidatus is a facultative symbiotic mollusc. In summary, although the data consistently suggest that N. vinctus is a non-symbiotic mollusc, inconsistencies in the δ13CBulk and δ15NBulk values of L. bicuspidatus make it difficult to determine whether this species has a symbiotic or a non-symbiotic lifestyle.
Benthic molluscs inhabiting organic rich sediments, such as the seafloor of an upwelling system2, are often deposit feeders77. However, the average δ13CBulk values for tissues of the bivalve L. bicuspidatus (− 16.7‰ ± 0.4‰) and the gastropod N. vinctus (− 17.5‰ ± 0.1‰) were higher (> 1.0‰ difference) than those of the sediments (− 21.1‰ to − 20.3‰). Thus, there is either a decoupling between molluscs and POM, or they are feeding selectively on some components of the bulk POC in the sediments that has a different δ13C value—possibly corresponding to diatoms and/or sulfur-oxidizing bacteria employing Rubisco Type II71. However, from our knowledge and from plotting spatial data from literature, sulfur bacteria microbial mats can be patchy and are either absent or low abundant (0.2 mm3 cm3) where the molluscs species are found (Supplementary Fig. S4). Generally 67% of the production derived from the perennial upwelling in the surface waters is assumed to arrive at 100 m depth of the Namibian shelf78, thus constantly replenishing the diatomaceous mud with fresh diatoms. The high δ13CBulk values of both species match the δ13CBulk values of diatoms in the BUS (− 18.0‰ to − 16.0‰)79. Moreover, the δ15NPhe values of L. bicuspidatus (4.7‰ ± 2.6‰) and N. vinctus (6.4‰ ± 0.2‰) are close to the δ15NPhe of diatoms’ consumers (5.3‰), namely pelagic mesozooplankton sampled at the central Namibian shelf by Steinkopf49. Together this suggests that for non-symbiotic organisms, biomass of freshly sedimented diatoms seems to be a more accessible nutritional source at the diatomaceous mud belt off Namibia, although bacteria may be ingested in an opportunistic way, either by microbes annexed to the diatoms agglutinations, episymbionts living in the gills, or when there is the opportunity to feed on bacterial mats.
Our TP values of L. bicuspidatus (1.7 ± 0.3) place this species between herbivory (TP of 2.0) and mixotrophy (TP of 1.5), again indicating that these animals are not incorporating bulk PON from the sediment but rather fresh material that still carries the unaltered autotrophic isotopic signature. The reasoning is that the TP of degraded PON can be expected to vary from 1.9 to 2.4 due to heterotrophic microbial processes63,80. Thus, the TP of L. bicuspidatus is too low for incorporation of degraded PON with an enhanced TP, supporting that animals obtained their nutrients from selective grazing of autotrophic material with a TP of 1.
The selective incorporation of autotrophic organisms by L. bicuspidatus may be by collecting fresh sedimented diatoms or free living bacteria with its labial palps77, or by nutrition from chemoautotrophic epibionts from the gills81. Interestingly, the TP range of L. bicuspidatus (1.4–1.9) was a surprisingly close fit to the TP range of the kleptoplastic gastropod P. ocellatus, which was 1.9 during algivory in the wild and 1.3 when reliant on chloroplasts for nutrition during lab-induced starvation67. Although there is no evidence yet of a symbiotic association between autotrophic bacteria and L. bicuspidatus, it is interesting to speculate that an undiscovered nutritional symbiosis with chemosynthetic bacteria could partially explain the low TP values of L. bicuspidatus tissue.
The TP of N. vinctus (2.6 ± 0.2) indicates an omnivorous lifestyle. At least 50% of the N. vinctus diet is from a heterotrophic source and the other 50% from an autotrophic source which, based on our δ13CBulk and δ15NPhe results, are likely to be diatoms. However, the δ15NPhe (6.4‰) of N. vinctus is slightly higher than that of herbivorous mesozooplankton from the central Namibian shelf (5.3‰)49 (Supplementary Table S2), which suggests that part of their diet comprises benthic autotrophs, i.e. heavier δ15N nitrate assimilating organisms at the base of the food web in the central Namibian shelf. Salt marshes mud-floating snails are known to feed on diatoms conspicuous at mud surface82,83, with the facultative carnivore Illyanassa obsolete84 potentially feeding upon annelids85. Similarly, N. vinctus has an extended foot that allows it to float at the surface of the diatoms mud. This probably allows it to feed on recently sedimented diatoms (δ15N from − 4.3 to 2.7‰) mixed with heavier δ15N nitrate (5.9‰ to 14.3‰) assimilating benthic chemoautotrophic bacteria (assumed to have a δ15N from 0.9 to 13.9‰ if a similar fractionation factor range for nitrate assimilation applies in all prokaryotes) (Supplementary Table S2) floating at the mud surface, while also supplementing its diet with carnivory (TP of 2.6 ± 0.2).
Ammonium cycling plays an important role in the dietary ecology of the deep water lucinid Lucinoma capensis
Sulfur-oxidising bacteria from the genus Ca. Thiodiazotropha are associated with lucinids from shallow-water habitats around the world and all Ca. Thiodiazotropha MAGs described to date encode the nif cluster of genes for fixing dinitrogen; a strategy critical for living in oligotrophic and nitrogen-limited ecosystems such as seagrass meadows and coral reefs10,11,12,13,14. However, none of the genes required for performing dinitrogen fixation were present in the MAGs of the Ca. Thiodiazotropha associated with L. capensis; this is unlikely to be a technical artefact as all three MAGs have a high level of completeness (> 99%). Furthermore, given that the δ15N of dissolved dinitrogen is 0.6‰ and the maximum isotope fractionation of dinitrogen fixation is 2.5‰50, the depleted δ15NBulk (− 4.9‰ ± 0.7‰) values in L. capensis gills are thus inconsistent with the symbionts obtaining nitrogen from dinitrogen fixation and point instead to alternative N-sources like nitrate or ammonium.
The δ15N nitrate in the sediment-overlying water in the central Namibian shelf ranges from 5.9 to 14.3‰51, and the isotopic fractionation of nitrate assimilation by prokaryotes from 0.4 to 5.0‰52. Accordingly, PON resulting from nitrate assimilation processes have δ15NBulk values varying from 0.9 to 13.9‰. Thus, the δ15NBulk (− 4.9‰ ± 0.7‰) values of the L. capensis gills remain lower than expected to conclude that nitrate is a significant source of nitrogen for the symbiosis. Additionally, our inability to detect genes for assimilatory nitrate reductases in the MAGs of the L. capensis symbionts further supports the exclusion of nitrate assimilation as the main process responsible for obtaining nitrogen for growth.
Assuming that the δ15N of ammonium largely reflects the δ15N of particulate organic nitrogen settling to the seafloor, and that δ15N fractionation during ammonification of PON in organic-rich marine sediments is negligible53,54,55, we can deduce the δ15N of the ammonium pool from sediment PON δ15N values, thus varying from 4.3 to 5.6‰ in the study area sediments (Supplementary Table S2). Taking into account an in situ ammonium concentration of 4 µM in the water collected from cores at Stn. 48 (Table 1) and that ammonium assimilation can have an isotope effect varying from 5 to 20‰ in marine microbes under an ambient ammonium concentration of approximately 4µM58, the δ15NBulk (− 4.9‰ ± 0.7‰) of L. capensis gills lead us to propose that the L. capensis symbionts rely on ammonium assimilation. This is clearly supported by the presence of the genes required for ammonium assimilation in the L. capensis symbiont MAGs (Table 2) and the threefold lower ammonium release rates in the L. capensis incubations compared to L. bicuspidatus and N. vinctus, a difference which is unlikely to be explained by a lower metabolic rate since oxygen consumption rates of L. capensis were similar to the bivalve L. bicuspidatus.
An alternative non-mutually exclusive possibility is that nitrogenous waste products excreted by L. capensis in the form of ammonium can be taken up and “internally recycled” by the symbionts24. Assuming that ammonium does not accumulate, in which case no isotope fractionation occurs86, “internal recycling” could easily explain the low δ15NBulk (− 4.9‰ ± 0.7‰) and possibly also the δ15NPhe (− 6.0‰ ± 1.8‰) values in the symbiotic gill tissue. Considering that the non-symbiotic tissue has a δ15NBulk value of -2.5‰, and the fractionation factor of ammonium excretion is 2.7‰87, the ammonium produced by the bivalve would have a δ15N of − 5.2‰ (Fig. 6). This value fits well to the bivalves gills δ15NBulk when considering that fractionation does not take place in closed systems when the reaction goes to completion86.
Schematic depiction of the ammonium assimilation and excretion pathways of Lucinoma capensis and its symbionts, based on the δ15N of different N pools. δ15NBulk, δ15NPhe and δ15NGlu were measured in this study (*). The δ15N of ammonium pool and ammonium excreted were calculated in this study based only on numbers provided in this scheme (**). We assumed that δ15N fractionation during ammonification of PON in organic-rich marine sediments is negligible53,54,55. The fractionation factor for ammonium assimilation is from Hoch et al.56. The fractionation factor for diet incorporation and internal ammonium assimilation in a closed system (recycling) are from Montoya86. The fractionation factor for ammonium excretion is from Checkley and Miller87. Figure made in Inkscape 1.1.2, www.inkscape.org.
It is important to highlight that an external source of nitrogen is still necessary for net growth88, and the L. capensis symbionts may be using both the environmental and host-excreted ammonium. The ammonium produced by L. capensis was low (0.14 µmol h−1 AFDW gram − 1 ± 0.1 (n = 3)) compared to the Mediterranean lucinid Loripes orbiculatus, which has a minimum average ammonium net excretion of about 5 µmol h−1 SFDW g−189. L. orbiculatus inhabits nitrogen-limited seagrass meadows in the Mediterranean where there is negligible ammonium available in the environment, so host and symbiont must thus rely on the thermodynamically expensive dinitrogen fixing process to meet their nitrogen requirements89. The BUS, on the other hand, is a productive upwelling region with high organic matter turnover, resulting in the formation of an OMZ that is rich in ammonium2. L. capensis can therefore exploit the high availability of ammonium in the reducing sediments of an OMZ as a reliable source of nitrogen39,90. These critical differences in the habitats of L. capensis and L. orbiculatus are likely to be key factors driving the differences in nitrogen metabolism seen between shallow and deeper lucinids’ symbionts.
The potential use of nitrate as an electron acceptor by an OMZ chemo-symbiotic mollusc
Nitrate consumed by L. capensis was at least threefold higher than that of both N. vinctus and L. bicuspidatus. Our findings further show that nitrate reduction is coupled with elevated nitrite net production, which is consistent with the activity of genes encoding the dissimilatory nitrate-reductase complex (narGHIJ) and the absence of the assimilatory pathway in the L. capensis symbiont MAGs (Table 2). These findings suggest that L. capensis symbionts use nitrate as an alternative electron acceptor90. Indeed, symbionts of L. aequizonata, which also inhabits an OMZ, are capable of nitrate respiration even under oxic conditions, and produce nitrite under aerobic conditions91. Nitrite net production rates in L. capensis incubations were highly variable and although we had only few replicates, they appeared to correlate with the oxygen concentration present during the incubation; the highest nitrite net production rate (0.3 µmol l−1 AFDW day−1) was observed in the vial with the highest oxygen concentrations during the whole incubation. The initial oxygen concentration in this vial was 3.23 ml l−1 and 2.1 ml l−1 at the end, while the other replicates reached anoxia at the end of their incubations. As genes encoding nitrite reductases were present in the L. capensis symbiont MAGs, this discrepancy may be explained by the inhibitory effects of high oxygen concentrations on nitrite reductase activity92. Future incubations with additional clam individuals or with physically enriched symbionts under defined conditions could be used to test this hypothesis.
Data availability
The metagenomic and amplicon sequencing data have been deposited with links to BioProject accession numbers PRJNA765502 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/). The BioSample accession numbers for the MAGs are SAMN21561986-SAMN21561988 and SAMN21572993-SAMN21572995 are for the corresponding raw read sets. All other datasets used during the current study are available from the corresponding author on reasonable request.
References
Schulz, H. N. et al. Dense populations of a giant sulfur bacterium in Namibian shelf sediments. Science 284, 493–495. https://doi.org/10.1126/science.284.5413.493%JScience (1999).
Brüchert, V. et al. Biogeochemical and physical control on shelf anoxia and water column hydrogen sulphide in the Benguela coastal upwelling system off Namibia. In Past and Present Water Column Anoxia (ed. Neretin, L. N.) 161–193 (Springer, 2006).
Currie, B., Utne-Palm, A. C. & Salvanes, A. G. V. Winning ways with hydrogen sulphide on the Namibian shelf. Front. Mar. Sci. 5, 341. https://doi.org/10.3389/fmars.2018.00341 (2018).
Emeis, K. C. et al. Shallow gas in shelf sediments of the Namibian coastal upwelling ecosystem. Cont. Shelf Res. 24, 627–642 (2004).
Eisenbarth, S. & Zettler, M. L. Diversity of the benthic macrofauna off northern Namibia from the shelf to the deep sea. J. Mar. Syst. 155, 1–10 (2016).
Zettler, M. L., Bochert, R. & Pollehne, F. Macrozoobenthos diversity in an oxygen minimum zone off northern Namibia. Mar. Biol. 156, 1949–1961. https://doi.org/10.1007/s00227-009-1227-9 (2009).
Cary, S. C., Vetter, R. D. & Felbeck, H. Habitat characterization and nutritional strategies of the endosymbiont-bearing bivalve Lucinoma aequizonata. Mar. Ecol. Prog. Ser. 55, 31–45 (1989).
Le Pennec, M., Beninger, P. G. & Herry, A. Feeding and digestive adaptations of bivalve molluscs to sulphide-rich habitats. Comp. Biochem. Physiol. A Physiol. 111, 183–189. https://doi.org/10.1016/0300-9629(94)00211-B (1995).
Taylor, J. D. & Glover, E. A. Functional anatomy, chemosymbiosis and evolution of the Lucinidae. Geol. Soc. Lond. Spec. Publ. 177, 207–225. https://doi.org/10.1144/GSL.SP.2000.177.01.12 (2000).
Lim, S. J. et al. Extensive thioautotrophic gill endosymbiont diversity within a single Ctena orbiculata (Bivalvia: Lucinidae) population and implications for defining host-symbiont specificity and species recognition. MSystems 4, e00280. https://doi.org/10.1128/mSystems.00280-19 (2019).
König, S. et al. Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat. Microbiol. 2, 16193. https://doi.org/10.1038/nmicrobiol.2016.193 (2016).
Petersen, J. M. et al. Chemosynthetic symbionts of marine invertebrate animals are capable of nitrogen fixation. Nat. Microbiol. 2, 16195. https://doi.org/10.1038/nmicrobiol.2016.195 (2016).
Osvatic, J. T. et al. Global biogeography of chemosynthetic symbionts reveals both localized and globally distributed symbiont groups. Proc. Natl. Acad. Sci. 118, e2104378118. https://doi.org/10.1073/pnas.2104378118 (2021).
Lim, S. J. et al. Taxonomic and functional heterogeneity of the gill microbiome in a symbiotic coastal mangrove lucinid species. ISME J. 13, 902–920. https://doi.org/10.1038/s41396-018-0318-3 (2019).
Taylor, J., Glover, E. & Williams, S. Diversification of chemosymbiotic bivalves: Origins and relationships of deeper water Lucinidae. Biol. J. Lin. Soc. 111, 401–420. https://doi.org/10.1111/bij.12208 (2014).
Taylor, J. & Glover, E. Biology, Evolution and Generic Review of the Chemosymbiotic Bivalve Family Lucinidae (Ray Society, 2021).
Nagel, B. et al. N-cycling and balancing of the N-deficit generated in the oxygen minimum zone over the Namibian shelf-An isotope-based approach. J. Geophys. Res. Biogeosci. 118, 361–371. https://doi.org/10.1002/jgrg.20040 (2013).
Neumann, A. & Flohr, A. The bivalve Lembulus bicuspidatus may enhance denitrification in shelf sediment at the Angola-Benguela Frontal Zone. Afr. J. Mar. Sci. 40, 91–96. https://doi.org/10.2989/1814232X.2018.1437774 (2018).
Sampaio, L., Rodrigues, A. M. & Quintino, V. Carbon and nitrogen stable isotopes in coastal benthic populations under multiple organic enrichment sources. Mar. Pollut. Bull. 60, 1790–1802. https://doi.org/10.1016/j.marpolbul.2010.06.003 (2010).
Sakko, A. L. The influence of the Benguela upwelling system on Namibia’s marine biodiversity. Biodivers. Conserv. 7, 419–433. https://doi.org/10.1023/A:1008867310010 (1998).
Levin, L. A., Mendoza, G. F., Konotchick, T. & Lee, R. Macrobenthos community structure and trophic relationships within active and inactive Pacific hydrothermal sediments. Deep Sea Res. II 56, 1632–1648. https://doi.org/10.1016/j.dsr2.2009.05.010 (2009).
Soto, L. A. Stable carbon and nitrogen isotopic signatures of fauna associated with the deep-sea hydrothermal vent system of Guaymas Basin, Gulf of California. Deep Sea Res. II 56, 1675–1682. https://doi.org/10.1016/j.dsr2.2009.05.013 (2009).
Weems, J., Iken, K., Gradinger, R. & Wooller, M. J. Carbon and nitrogen assimilation in the Bering Sea clams Nuculana radiata and Macoma moesta. J. Exp. Mar. Biol. Ecol. 430, 32–42. https://doi.org/10.1016/j.jembe.2012.06.015 (2012).
Ferrier-Pagès, C. & Leal, M. C. Stable isotopes as tracers of trophic interactions in marine mutualistic symbioses. Ecol. Evol. 9, 723–740. https://doi.org/10.1002/ece3.4712 (2019).
DavySimon, K., Allemand, D. & WeisVirginia, M. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. https://doi.org/10.1128/MMBR.05014-11 (2012).
Ferrier-Pagès, C. et al. Summer autotrophy and winter heterotrophy in the temperate symbiotic coral Cladocora caespitosa. Limnol. Oceanogr. 56, 1429–1438. https://doi.org/10.4319/lo.2011.56.4.1429 (2011).
Berg, C. J. & Alatalo, P. Potential of chemosynthesis in molluscan mariculture. Aquaculture 39, 165–179. https://doi.org/10.1016/0044-8486(84)90264-3 (1984).
Dando, P. R. & Southward, A. J. Chemoautotrophy in bivalve molluscs of the genus Thyasira. J. Mar. Biol. Assoc. U.K. 66, 915–929. https://doi.org/10.1017/S0025315400048529 (1986).
Spiro, B., Greenwood, P. B., Southward, A. J. & Dando, P. R. 13C/12C ratios in marine invertebrates from reducing sediments: Confirmation of nutritional importance of chemoautotrophic endosymbiotic bacteria. Mar. Ecol. Prog. Ser. 28, 233–240 (1986).
Fisher, C. R. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. Rev. Aquat. Sci. 2, 399–436 (1990).
Duperron, S., Fiala-Medioni, A., Caprais, J. C., Olu, K. & Sibuet, M. Evidence for chemoautotrophic symbiosis in a Mediterranean cold seep clam (Bivalvia: Lucinidae): Comparative sequence analysis of bacterial 16S rRNA, APS reductase and RubisCO genes. FEMS Microbiol. Ecol. 59, 64–70. https://doi.org/10.1111/j.1574-6941.2006.00194.x (2007).
Zanzerl, H., Salvo, F., Jones, S. W. & Dufour, S. C. Feeding strategies in symbiotic and asymbiotic thyasirid bivalves. J. Sea Res. 145, 16–23. https://doi.org/10.1016/j.seares.2018.12.005 (2019).
Descolas-Gros, C. & Fontugne, M. R. Carbon fixation in marine phytoplankton: Carboxylase activities and stable carbon-isotope ratios; physiological and paleoclimatological aspects. Mar. Biol. 87, 1–6. https://doi.org/10.1007/BF00396999 (1985).
Brooks, J. M. et al. Deep-sea hydrocarbon seep communities: Evidence for energy and nutritional carbon sources. Science 238, 1138. https://doi.org/10.1126/science.238.4830.1138 (1987).
Conway, N., Capuzzo, J. M. & Fry, B. The role of endosymbiotic bacteria in the nutrition of Solemya velum: Evidence from a stable isotope analysis of endosymbionts and host. Limnol. Oceanogr. 34, 249–255. https://doi.org/10.4319/lo.1989.34.1.0249 (1989).
Conway, N. M., Howes, B. L., McDowell Capuzzo, J. E., Turner, R. D. & Cavanaugh, C. M. Characterization and site description of Solemya borealis (Bivalvia; Solemyidae), another bivalve-bacteria symbiosis. Mar. Biol. 112, 601–613. https://doi.org/10.1007/BF00346178 (1992).
Rau, G. H. Low 15N/14N in hydrothermal vent animals: Ecological implications. Nature 289, 484. https://doi.org/10.1038/289484a0 (1981).
Kennicutt, M. C. et al. Stable isotope partitioning in seep and vent organisms: Chemical and ecological significance. Chem. Geol. Isot. Geosci. Sect. 101, 293–310. https://doi.org/10.1016/0009-2541(92)90009-T (1992).
Lee, R. W. & Childress, J. J. Assimilation of inorganic nitrogen by marine invertebrates and their chemoautotrophic and methanotrophic symbionts. Appl. Environ. Microbiol. 60, 1852–1858. https://doi.org/10.1128/AEM.60.6.1852-1858.1994 (1994).
Minagawa, M. & Wada, E. Stepwise enrichment of 15N along food chains: Further evidence and the relation between δ15N and animal age. Geochim. Cosmochim. Acta 48, 1135–1140. https://doi.org/10.1016/0016-7037(84)90204-7 (1984).
Zanden, M. J. V. & Rasmussen, J. B. Variation in δ15N and δ13C trophic fractionation: Implications for aquatic food web studies. Limnol. Oceanogr. 46, 2061–2066. https://doi.org/10.4319/lo.2001.46.8.2061 (2001).
Nagel, B., Gaye, B., Lahajnar, N., Struck, U. & Emeis, K.-C. Effects of current regimes and oxygenation on particulate matter preservation on the Namibian shelf: Insights from amino acid biogeochemistry. Mar. Chem. 186, 121–132. https://doi.org/10.1016/j.marchem.2016.09.001 (2016).
Holmes, M. E. et al. Stable nitrogen isotopes in Angola Basin surface sediments. Mar. Geol. 134, 1–12. https://doi.org/10.1016/0025-3227(96)00031-X (1996).
Post, D. M. Using stable isotopes to estimate trophic position: Models, methods, and assumptions. Ecology 83, 703–718. https://doi.org/10.1890/0012-9658(2002)083[0703:USITET]2.0.CO;2 (2002).
McClelland, J. W. & Montoya, J. P. Trophic relationships and the nitrogen isotopic composition of amino acids in plankton. Ecology 83, 2173–2180 (2002).
Chikaraishi, Y. et al. Determination of aquatic food-web structure based on compound-specific nitrogen isotopic composition of amino acids. Limnol. Oceanogr. Methods 7, 740–750. https://doi.org/10.4319/lom.2009.7.740 (2009).
Glibert, P. M., Middelburg, J. J., McClelland, J. W. & Jake Vander Zanden, M. Stable isotope tracers: Enriching our perspectives and questions on sources, fates, rates, and pathways of major elements in aquatic systems. Limnol. Oceanogr. 64, 950–981. https://doi.org/10.1002/lno.11087 (2019).
Mompeán, C., Bode, A., Gier, E. & McCarthy, M. D. Bulk vs amino acid stable N isotope estimations of metabolic status and contributions of nitrogen fixation to size-fractionated zooplankton biomass in the subtropical N Atlantic. Deep Sea Res. I 114, 137–148. https://doi.org/10.1016/j.dsr.2016.05.005 (2016).
Steinkopf, M. Trophische Strukturen des Mesozooplanktons im Benguela Auftriebsgebiet vor Namibia (Universität Rostock, 2018).
Sigman, D. & Fripiat, F. Nitrogen isotopes in the Ocean. In Encyclopedia of Ocean Sciences 3rd edn, Vol. 263 (eds Cochran, J. K. et al.) 268 (Academic Press, 2019).
Nagel, B. et al. Nutrients and δ15N measured in water samples in the oxygen minimum zone over the Namibian shelf during the Meteor campaign M76–2 in 2008. PANGAEA. https://doi.org/10.1594/PANGAEA.892369 (2018).
Granger, J., Sigman, D. M., Rohde, M. M., Maldonado, M. T. & Tortell, P. D. N and O isotope effects during nitrate assimilation by unicellular prokaryotic and eukaryotic plankton cultures. Geochim. Cosmochim. Acta 74, 1030–1040 (2010).
Prokopenko, M. G., Hammond, D. E. & Stott, L. Lack of isotopic fractionation of δ 15N of organic matter during long-term diagenesis in marine sediments, ODP Leg 202, Sites 1234 and 1235. In Proc. Ocean Drilling Program(eds. R. Tiedemann, A. C. Mix, C. Richter and W. F. Ruddiman) 22 (2006).
Prokopenko, M. G. et al. Nitrogen cycling in the sediments of Santa Barbara basin and Eastern Subtropical North Pacific: Nitrogen isotopes, diagenesis and possible chemosymbiosis between two lithotrophs (Thioploca and Anammox)—“Riding on a glider”. Earth Planet. Sci. Lett. 242, 186–204 (2006).
Robinson, R. S. et al. A review of nitrogen isotopic alteration in marine sediments. Paleoceanography 27, 4203. https://doi.org/10.1029/2012PA002321 (2012).
Hoch, M. P., Fogel, M. L. & Kirchman, D. L. Isotope fractionation during ammonium uptake by marine microbial assemblages. Geomicrobiol. J. 12, 113–127. https://doi.org/10.1080/01490459409377977 (1994).
Grasshoff, K. et al. (eds) Methods of Seawater Analysis 3rd edn. (Wiley, 2009).
Hofmann, D., Gehre, M. & Jung, K. Sample preparation techniques for the determination of natural 15N/14N variations in amino acids by gas chromatography-combustion-isotope ratio mass spectrometry (GC-C-IRMS). Isot. Environ. Health Stud. 39, 233–244. https://doi.org/10.1080/1025601031000147630 (2003).
Veuger, B., Middelburg, J. J., Boschker, H. T. S. & Houtekamer, M. Analysis of 15N incorporation into D-alanine: A new method for tracing nitrogen uptake by bacteria. Limnol. Oceanogr. Methods 3, 230–240. https://doi.org/10.4319/lom.2005.3.230 (2005).
Loick-Wilde, N. et al. Stratification, nitrogen fixation, and cyanobacterial bloom stage regulate the planktonic food web structure. Glob. Change Biol. 25, 794–810. https://doi.org/10.1111/gcb.14546 (2019).
Chikaraishi, Y., Ogawa, N. O., Doi, H. & Ohkouchi, N. 15N/14N ratios of amino acids as a tool for studying terrestrial food webs: A case study of terrestrial insects (bees, wasps, and hornets). Ecol. Res. 26, 835–844. https://doi.org/10.1007/s11284-011-0844-1 (2011).
Chikaraishi, Y. et al. High-resolution food webs based on nitrogen isotopic composition of amino acids. Ecol. Evol. 4, 2423–2449. https://doi.org/10.1002/ece3.1103 (2014).
Eglite, E. et al. Strategies of amino acid supply in mesozooplankton during cyanobacteria blooms: A stable nitrogen isotope approach. Ecosphere 9, e02135. https://doi.org/10.1002/ecs2.2135 (2018).
Fujii, T. et al. Organic carbon and nitrogen isoscapes of reef corals and algal symbionts: Relative influences of environmental gradients and heterotrophy. Microorganisms 8, 1221. https://doi.org/10.3390/microorganisms8081221 (2020).
Ferrier-Pagès, C. et al. Tracing the trophic plasticity of the coral–dinoflagellate symbiosis using amino acid compound-specific stable isotope analysis. Microorganisms 9, 182. https://doi.org/10.3390/microorganisms9010182 (2021).
Hannides, C. C. S., Popp, B. N., Landry, M. R. & Graham, B. S. Quantification of zooplankton trophic position in the North Pacific Subtropical Gyre using stable nitrogen isotopes. Limnol. Oceanogr. 54, 50–61. https://doi.org/10.4319/lo.2009.54.1.0050 (2009).
Maeda, T. et al. Algivore or phototroph? Plakobranchus ocellatus (Gastropoda) continuously acquires kleptoplasts and nutrition from multiple algal species in nature. PLoS ONE 7, e42024. https://doi.org/10.1371/journal.pone.0042024 (2012).
Pjevac, P. et al. An economical and flexible dual barcoding, two-step PCR approach for highly multiplexed amplicon sequencing. Front. Microbiol. 12, 1069 (2021).
Brettin, T. et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. https://doi.org/10.1038/srep08365 (2015).
Steffan, S. A. et al. Unpacking brown food-webs: Animal trophic identity reflects rampant microbivory. Ecol. Evol. 7, 3532–3541. https://doi.org/10.1002/ece3.2951 (2017).
Robinson, J. J. & Cavanaugh, C. M. Expression of form I and form II Rubisco in chemoautotrophic symbioses: Implications for the interpretation of stable carbon isotope values. Limnol. Oceanogr. 40, 1496–1502. https://doi.org/10.4319/lo.1995.40.8.1496 (1995).
Fry, B. Stable Isotope Ecology (Springer, 2006).
Emeis, K. et al. pCO2 underway data from the Benguela upwelling system in southeastern South Atlantic Ocean. PANGAEA. https://doi.org/10.1594/PANGAEA.880406 (2017).
Mook, W. G., Bommerson, J. C. & Staverman, W. H. Carbon isotope fractionation between dissolved bicarbonate and gaseous carbon dioxide. Earth Planet. Sci. Lett. 22, 169–176 (1974).
Goericke, R., Montoya, J. & Fry, B. Physiology and isotopic fractionation in algae and cyanobacteria. In Stable Isotopes in Ecology and Environmental Science (eds Kajtah, K. & Michener, R. H.) 187–221 (Blackwell, 1994).
Duplessis, M. R., Dufour, S. C., Blankenship, L. E., Felbeck, H. & Yayanos, A. A. Anatomical and experimental evidence for particulate feeding in Lucinoma aequizonata and Parvilucina tenuisculpta (Bivalvia: Lucinidae) from the Santa Barbara Basin. Mar. Biol. 145, 551–561. https://doi.org/10.1007/s00227-004-1350-6 (2004).
Lopez, G. R. & Levinton, J. S. Ecology of deposit-feeding animals in marine Sediments. Q. Rev. Biol. 62, 235–260. https://doi.org/10.1086/415511 (1987).
Brüchert, V. et al. Regulation of bacterial sulfate reduction and hydrogen sulfide fluxes in the central Namibian coastal upwelling zone. Geochim. Cosmochim. Acta 67, 4505–4518 (2003).
Schukat, A., Auel, H., Teuber, L., Lahajnar, N. & Hagen, W. Complex trophic interactions of calanoid copepods in the Benguela upwelling system. J. Sea Res. 85, 186–196. https://doi.org/10.1016/j.seares.2013.04.018 (2014).
McCarthy, M. D., Benner, R., Lee, C. & Fogel, M. L. Amino acid nitrogen isotopic fractionation patterns as indicators of heterotrophy in plankton, particulate, and dissolved organic matter. Geochim. Cosmochim. Acta 71, 4727–4744. https://doi.org/10.1016/j.gca.2007.06.061 (2007).
Zbinden, M. et al. Epsilonproteobacteria as gill epibionts of the hydrothermal vent gastropod Cyathermia naticoides (North East-Pacific Rise). Mar. Biol. 162, 435–448. https://doi.org/10.1007/s00227-014-2591-7 (2015).
Whitlatch, R. B. & Obrebski, S. Feeding selectivity and coexistence in two deposit-feeding gastropods. Mar. Biol. 58, 219–225. https://doi.org/10.1007/BF00391879 (1980).
Connor, M. S. & Robert, K. E. Selective grazing by the mud snail Ilyanassa obsoleta. Oecologia 53, 271–275 (1982).
Feller, R. J. Dietary immunoassay of Ilyanassa obsoleta, the eastern mud snail. Biol. Bull. 166, 96–102. https://doi.org/10.2307/1541433 (1984).
Kelaher, B. P., Levinton, J. S. & Matthew Hoch, J. Foraging by the mud snail, Ilyanassa obsoleta (Say), modulates spatial variation in benthic community structure. J. Exp. Mar. Biol. Ecol. 292, 139–157. https://doi.org/10.1016/S0022-0981(03)00183-7 (2003).
Montoya, J. P. Natural abundance of 15N in marine planktonic ecosystems. In Stable Isotopes in Ecology and Environmental Science (eds Michener, R. & Lajtha, K.) 176–201 (Blackwell Publishing Ltd, 2007).
Checkley, D. M. & Miller, C. A. Nitrogen isotope fractionation by oceanic zooplankton. Deep Sea Res. A Oceanogr. Res. Pap. 36, 1449–1456. https://doi.org/10.1016/0198-0149(89)90050-2 (1989).
Nelson, D. C. & Fisher, C. R. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps. In The Microbiology of Deep-Sea Hydrothermal Vents (ed. Karl, D. M.) 125–167 (CRC Press, 1995).
Cardini, U. et al. Chemosymbiotic bivalves contribute to the nitrogen budget of seagrass ecosystems. ISME J. 13, 3131–3134. https://doi.org/10.1038/s41396-019-0486-9 (2019).
Lee, R. W., Robinson, J. J. & Cavanaugh, C. M. Pathways of inorganic nitrogen assimilation in chemoautotrophic bacteria-marine invertebrate symbioses: Expression of host and symbiont glutamine synthetase. J. Exp. Biol. 202, 289 (1999).
Hentschel, U. & Felbeck, H. Nitrate respiration in chemoautotrophic symbionts of the bivalve Lucinoma aequizonata is not regulated by oxygen. Appl. Environ. Microbiol. 61, 1630–1633 (1995).
Sacks, L. E. & Barker, H. A. The influence of oxygen on nitrate and nitrite reduction. J. Bacteriol. 58, 11–22. https://doi.org/10.1128/JB.58.1.11-22.1949 (1949).
Acknowledgements
The authors are grateful to Christian Burmeister (Leibniz-Institute for Baltic Sea Research), who handled the auto-analyser (QuAAtro, Seal analytical), to Iris Liskow (Leibniz-Institute for Baltic Sea Research) who handled the Elemental Analyser (EA, Thermo Flash EA 1112), and to Markus Steinkopf and Ina Schmidt (Leibniz-Institute for Baltic Sea Research), who carried out the laboratory work with the compound-specific isotope analyses (CSIA) of amino acid nitrogen. We are also thankful to Prof. Dr. Heide Schulz-Vogt for her critical look at our nutrient measurements results and contribution with information about large sulfur bacteria abundance in the Namibian shelf. We are grateful to Dr. Andy Dale for his critical look at our benthic mineralization assumptions. We also would like to thank the captain and crew of RV Meteor for the assistance during the cruise sampling. The present study was part of a project EVAR funded by the Federal Ministry of Education and Research (Grant No.: 03V01279).
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
K.A. designed the study; M.L.Z., K.A, N.L.W., B.Y., J.M.P., J.T.O, J.W.R., J.F contributed to theoretical concepts applied in the study; K.A. carried out the experiments, M.L.Z., K.A, J.W.R, J.F collected the samples; K.A., B.Y., D.W. processed the samples; K.A., N.L.W., B.Y., J.T.O., B.H. analysed the data; K.A. wrote the article; K.A., N.L.W., B.Y., M.L.Z, contributed significantly to the discussion; K.A., N.L.W., B.Y., J.F, M.L.Z, contributed significantly to the revision of the paper; all the authors contributed significantly for the final text.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Amorim, K., Loick-Wilde, N., Yuen, B. et al. Chemoautotrophy, symbiosis and sedimented diatoms support high biomass of benthic molluscs in the Namibian shelf. Sci Rep 12, 9731 (2022). https://doi.org/10.1038/s41598-022-13571-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-13571-w