Among the many wonders that microbes can achieve, two fundamental aspects make the microbial communities successful in conquering all the imaginable environments on Earth. One is their metabolic versatility. For any substrate that exists on our planet, there might be a few bacterial species able to use it for energy and growth. The ability to use any types of substrates often relies on how multiple metabolisms assemble together. In fact, the second striking aspect is microbes' ability to organize themselves in structured communities. Their spatial arrangement is the key to maintaining their metabolic diversity (1,2). However, these structured communities live in an ever-changing environment. One can easily imagine plentiful examples in which the local conditions of the microbial communities change. After the rain, the soil goes from dry to wet; in our stomach, sudden new nutrients are available after every meal; intertidal zones change continuously cycling between tides, and so forth. The reason behind our work was precisely to understand the mechanisms that help maintain microbial diversity during range expansion while the surrounding environment is changing.

When I joined David Johnson's group, he was working with his former Ph.D., Felix Goldschmidt on crucial aspects that govern spatial diversity in range expanding microbial communities. This model system enables to study the fundamental principles of ecology underlying the population's spatial dynamic (3-9). When a population of bacteria land on a pristine agar surface, interactions with neighbors, racing for space and nutrient, contribute to the emergence of the spatial arrangement of the community as a whole. The outcome of these interactions is driven by the initial spatial position and the nature of interactions cells engage during its expansion (10). Besides the biotic factors, environmental conditions play an important role in shaping these interactions (1,2). However, the extent of the contribution of the latest to the spatial development of microbial communities is not yet understood (1,11). Often, the main problem is the complexity of the natural environment; therefore, controlled laboratory conditions and ad hoc designed synthetic bacterial consortia come in handy (11).
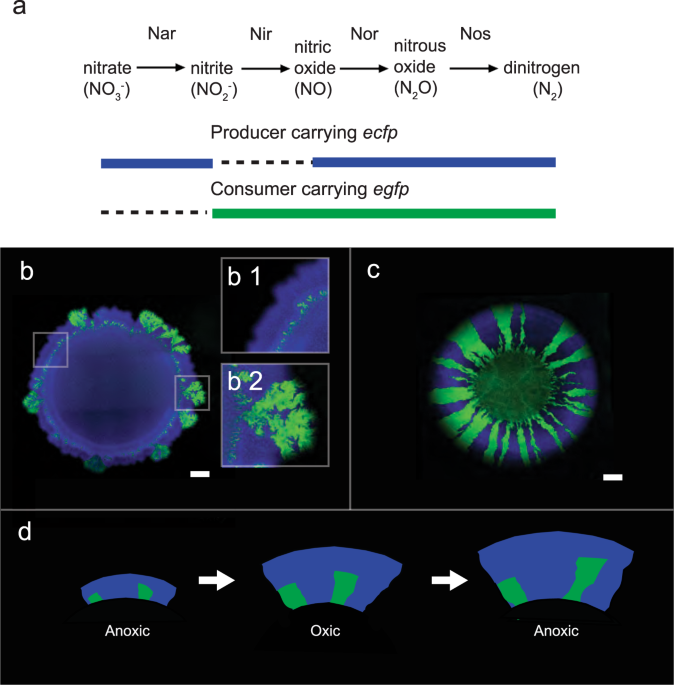
Our model system uses two metabolically dependent knockouts of P. stutzeri that exchange nitrite as an intermediate metabolite in the denitrification pathway. One converts nitrate to nitrite (producer) and the other reduces nitrite to nitrogen gas (consumer). Felix's work established that our consortium incubated at two different pH in continuous anaerobic conditions would lead to two distinct patterns and diversity (10). At pH 7.5, the producer expands first, leaving behind the consumer. In contrast, at pH 6.5, nitrite toxicity imposes stronger mutualistic interaction that promotes a concurrent expansion of the consumer and producer (12,13). Their insights (10,12,13) were fundamental to building a solid ground for this research. While they established that the two above mentioned patterns (producer first and concurrent) would emerge under anaerobic conditions, we were curious whether incubating the same plate under fluctuating conditions between aerobic and anaerobic conditions would impact the diversity and lead to the loss of consumers. We argued that if the anaerobic conditions lead to a loss of consumers, particularly at pH 7.5; incubating plates in aerobic conditions afterward would result in the complete loss of consumers. If this hypothesis had been true, a specialized metabolism like the consumer would disappear anywhere one can observe fluctuating conditions.
With this hypothesis in mind, we designed an experiment where we incubated plates for 24h under anaerobic conditions and afterward 12h under aerobic conditions. We kept cycling back and forth from anaerobic to aerobic conditions for a total of 350h. At each transition, I acquired the image of each colonies manually with a confocal laser scanning microscope. We used the images to quantify and compare the spatial evolution of the communities incubated on plates at pH 6.5, with those growing at pH 7.5. We discovered a persistent pattern where the consumers decreased significantly in their abundance at pH 7.5 when compared to pH 6.5 – but the underlying mechanisms that cause such a loss in diversity were not yet evident. Together with Gabriele Micali we worked on a script to dive into the images and extract information about the colonies' spatial organization. We designed a spatial analysis that quantified the local mixing and the overall surface of consumer to producer ratio over time. These rather simple measures enabled us to unveil something more complex like the two communities' spatiotemporal dynamic and finally narrow down the moment in which the two communities diverge from each other. Specifically, we pinpointed when at pH 7.5 the colonies reached a stable and lower spatial diversity than those at pH 6.5. Nevertheless, in both communities, consumers persisted.

Behind the scene: dissecting the Jackpot events. a) A sketch I used to discuss the dynamic of change in the local spatial arrangement of the consumer within the jackpot event when transitioning between anaerobic to aerobic condition. b-c) Early attempts to track individual jackpot event and capture the changing spatial arrangement over each transition in a fluctuating redox state.
Now we were able to get to the core of this project, the conversations with Dani Or and Dave spiced up. We looked into new directions and explored further aspects that were hiding in plain sight. We observed that the two communities, over time, decreased their spatial diversity. However, the strong mutualistic communities (pH 6.5) exhibit higher spatial diversity compared to the weak mutualistic ones (pH 7.5). A common aspect was that rare structures emerged in both communities that are often described as jackpot events in the form of large branches consisting solely of consumer cells. These structures usually arise when a mutation occurs; however, the mutation argument was already ruled out (10,12,13). Therefore, these structures were purely rare events driven by other processes. Dani Or argued for such a structure to emerge and persist under a fluctuating environment, there should have been a mechanistic explanation that we did not have yet. We suspected that the answer was something we couldn't see or quantify simply using our microscopy images.

Hidden information within the image. During each microscopy acquisition, multiple type of images were recorded, besides the fluorescence channels to distinguish the two metabolic interacting strains (egfp for the consumer, and ecfp for the producer) also the reflective light and bright field were acquired. In addition, the latest unveiled that within the biomass, each transition between anaerobic to aerobic conditions was literally recorded. This observation, helped us to reason on the effect of each redox change on the spatial arrangement of the jackpot events.
At the time, I was commuting almost daily with Benedict Borer from Basel to Zurich. We discussed the possibility of creating an agent-based model to answer Dani Or’s questions. Benedict absorbed every word we exchanged and translated it into an elegant mathematical model that unveiled something we could not reveal with the experimental results. A metabolic landscape was hiding beneath our colonies. This finding was novel to us, so much that we published the research (14) which, together with a previous publication from Felix (10), became essential to set the path to answer Dani Or's questions for this current research. After several simulations, a reasonable amount of coffees, and a staggering number of commuting hours, we finally had the answers we were looking for. The emergence of the jackpot event was driven by local nutrient niches that result from the spatial metabolic landscape. In short, the localized metabolic interactions between the two dependent strains shaped how metabolites were accumulating at the micro scale. Furthermore, each pH condition was driving a different growth rate at each colony's expanding edge, creating conditions that were detrimental specifically for the consumers, leading to their loss, at pH 7.5. In contrast, at pH 6.5 the consumer's jackpot would persist better.
This work provides a simple approach to studying the influence of fluctuating environments between anaerobic and aerobic conditions on the spatial self-organization of bacterial communities. Most importantly, we showed that the spatial diversity that emerges in such situations is a consequence of the strength of metabolic interactions. We provided evidence of the critical role of the bacterial spatial organization as a key element for the microbial communities to persist and maintain their diversity in a fluctuating world.
References
- Ciccarese D, Johnson DR. Functional microbial landscapes. In: Moo-Young M (ed).579 Comprehensive Biotechnology. 3rd edn. (Elsevier, Amsterdam, 2019) vol 6, pp 42-51
- Ciccarese D, Zuidema A, Merlo V, Johnson DR. Interaction-dependent effects of surface structure on microbial spatial self-organization. Philos Trans R Soc Lond B Biol Sci. 2020;375:20190246
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci U S A. 2007;104:19926-30.
- Hallatschek O, Nelson DR. Population genetics and range expansions. Phys Today. 2009;62:42-7.
- Müller MJI, Neugeboren BI, Nelson DR, Murray AW. Genetic drift opposes mutualism during spatial population expansion. Proc. Natl Acad. Sci. USA. 2014;111:1037–1042
- van Gestel J, Weissing FJ, Kuipers OP, Kovács AT. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 2014;8:2069–2079
- Mitri S, Clarke E, Foster KR. Resource limitation drives spatial organization in microbial groups. ISME J. 2015;10:1471–1482
- Tecon R, Or D. Cooperation in carbon source degradation shapes spatial self-organization of microbial consortia on hydrated surfaces. Sci. Rep. 2017;7:43726.
- Giometto A, Nelson DR, Murray AW. Physical interactions reduce the power of natural selection in growing yeast colonies. Proc. Natl Acad. Sci. 2018;115:11448–11453
- Goldschmidt F, Caduff L, Johnson DR. Causes and consequences of pattern diversification in a spatially self-organizing microbial community. ISME J. 2021;8:2415-2426
- Tecon R, Mitri S, Ciccarese D, Or D, van der Meer JR, Johnson DR. Bridging the holistic-reductionist divide in microbial ecology. mSystems. 2019;4:e00265-18
- Goldschmidt F, Regoes RR, Johnson DR. Metabolite toxicity slows local diversity loss during expansion of a microbial cross-feeding community. ISME J. 2018;12:136-44
- Goldschmidt F, Regoes RR, Johnson DR. Successive range expansion promotes diversity and accelerates evolution in spatially structured microbial populations. ISMEJ. 2017;11:2112-23
- Borer B, Ciccarese D, Johnson DR, Or D. Spatial organization in microbial range expansion emerging from trophic dependencies and successful lineages. Commun Biol. 2020;3:685.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in