Infection with the SARS-CoV-2 virus can lead to diverse outcomes, ranging from no symptoms to varying degrees of disease severity, spanning mild illness to death. What determines the degree of severity is unclear, but mounting evidence points to exacerbated and abnormal responses in the innate branch of the immune system as a main driver of major illness. Writing in Nature, Combes et al.1 present a study investigating the hallmarks of COVID-19 severity.
Read the paper: Global absence and targeting of protective immune states in severe COVID-19
The authors analysed cells, including immune cells, in blood samples from 21 people with COVID-19 and 25 uninfected individuals who were either healthy or had a lung injury or breathing difficulties. They monitored gene expression during the course of the infection as patients went on to develop either what was categorized as mild–moderate COVID-19 (which required a short hospital stay without the need for intensive care or mechanical ventilation) or severe COVID-19 (requiring intensive care and mechanical ventilation). The authors found that the cells of people with mild–moderate COVID-19 expressed a distinct set of genes whose expression depends on what are known as type I interferon proteins. Interferons, molecules that are also called cytokines, drive the expression of genes that have a role in antiviral defence.
This interferon-regulated gene-expression signature was not observed in the cells of people with severe COVID-19. Instead, the cells had a gene-expression signature described as an inflammatory S100A12 myeloid-cell program (S100A12 is a protein expressed as part of this program). A S100A12 signature was previously identified2 as being associated with severe COVID-19. Interestingly, a similar program is associated with another form of severe disease called sepsis, which derives from an aberrant immune response to bacterial infection3.
An interferon-regulated gene-expression program can be crucial to defence against viral infection, so the lack of activation of this program in people with severe COVID-19 provided a hint that defective initiation of this pathway might contribute to the observed differences in disease severity. Combes et al. therefore set out to determine the reason for the differences. The first obvious suspect was the level of an interferon protein (IFN-α) in blood plasma (blood lacking its cellular content). The authors found no notable difference in IFN-α levels with differing disease severity. However, there are other types of interferon protein that the authors did not measure.
The authors next turned their attention to antibodies. Antibodies against SARS-CoV-2 have a protective role in the natural immune response to this virus, and antibodies targeting the virus have been used as COVID-19 treatments. Indeed, part of the rationale for using the vaccines currently available is to drive the generation of such antibodies. The authors found that the level of antibodies against the SARS-CoV-2 spike and nucleocapsid proteins was higher in people with severe disease than in those with mild–moderate COVID-19. Moreover, high antibody levels were negatively correlated with the presence of cells expressing an interferon-regulated gene-expression program.
Interferon deficiency can lead to severe COVID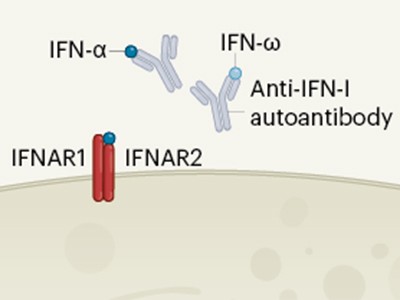
To search for a missing link between antibody levels, interferon and COVID-19 severity, Combes and colleagues used an in vitro system. They took immune cells from the blood of healthy people, and exposed them to blood plasma samples from people with mild–moderate or severe COVID-19. The authors then stimulated the immune cells with IFN-α, to determine whether an antiviral response developed. They found that the presence of plasma from people with severe disease blocked the induction of interferon-responsive genes. However, if this plasma was treated to deplete it of antibodies, interferon-mediated gene expression was restored in these immune cells.
Intriguingly, in a previous study4 of 987 individuals with severe COVID-19, 135 (13.7%) had anti-interferon antibodies that could blunt the induction of interferon-induced genes. However, given that most individuals in that study did not have anti-interferon antibodies, the presence of such antibodies alone could not fully explain the development of severe COVID-19. Indeed, Combes et al. found anti-interferon antibodies at a similarly low frequency in the samples they had obtained from patients.
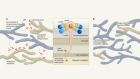
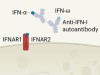
In an attempt to explain the enigma of a dampened interferon response in severe COVID-19, the authors considered various aspects of antibody function. An antibody consists structurally of two functional units: a variable region that recognizes the disease-causing agent and a constant region (termed Fc) that engages Fc receptors on the surface of immune cells (Fig. 1). This latter interaction can help to shape the immune response. During the course of a disease, the characteristics of the antibodies produced change to regulate immune defences. One aspect of these changes is an alteration in the antibody Fc component that affects which Fc receptors are engaged. For example, engagement with the Fc receptors CD64, CD16 and CD32 can regulate how the immune system eliminates bacterial and viral infections5.
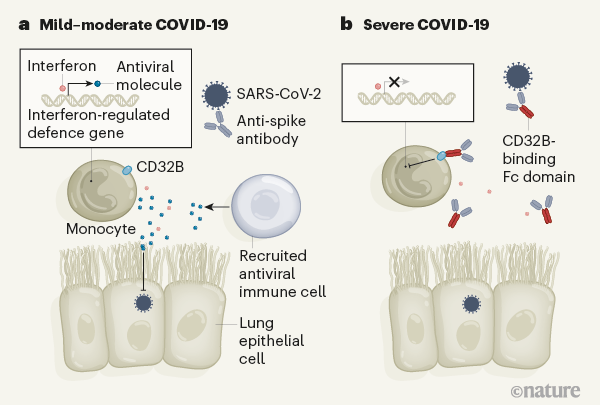
Figure 1 | Antibodies that affect immune defence in severe COVID-19. Combes et al.1 analysed blood samples from people who had COVID-19 of differing degrees of severity. a, In what the authors classify as mild–moderate cases of this illness, patients made antibodies that recognize the spike protein of the SARS-CoV-2 virus. The authors report that immune cells called monocytes from these individuals express genes that are regulated by the protein interferon. These genes encode molecules that aid antiviral defence by boosting immune responses or by suppressing the virus at sites of infection, which might include the epithelial cells that line the lungs. The CD32B receptor, which is found on monocytes, can dampen such immune responses if it is bound by another protein. b, The authors report that individuals with severe COVID-19 have anti-spike antibodies in which a region of the antibody, called the Fc domain, binds to CD32B. This interaction hinders the expression of interferon-regulated genes.
Combes and colleagues investigated whether Fc-receptor engagement has a role in blunting interferon-mediated responses in severe COVID-19. Using immune cells from healthy donors exposed to IFN-α and plasma from people with severe COVID-19, they individually blocked CD64, CD16 and CD32 Fc receptors. Only CD32 blockade enabled the expression of interferon-regulated genes.
The CD32 Fc receptor exists in two forms — CD32A and CD32B. CD32A engagement activates the immune system, whereas CD32B dampens immune responses6. Combes and colleagues showed that the inhibition of interferon-regulated gene expression was due to CD32B. They thus conclude that people with severe COVID-19 develop antibodies that engage with CD32B Fc receptors and thereby blunt interferon-mediated defence responses.
In support of this conclusion, a previous study7 demonstrated that people with moderate and severe COVID-19 develop a diverse antibody response in terms of the Fc regions recruited, with the presence of spike-specific antibodies that engage CD32B being a main predictor of disease severity. What determines this difference in antibody type between severe and moderate COVID-19 remains to be discovered.
Signs of self-sustained inflammatory circuits in severe COVID pneumonia
It is tempting to speculate that changing the Fc domain to one that engages CD32B is a mechanism used by the immune system to shut down an intense immune response to SARS-CoV-2. It would be interesting to investigate whether such mechanisms are involved in other types of viral infection, and, if so, whether they have a detrimental or beneficial role. Of note, there are also reported examples of SARS-CoV-2 infection generating antibodies that turn against the host. People with COVID-19 can develop antibodies that target nucleic acids8 and host proteins9.
It is important to remember that we do not yet know whether, in people with severe COVID-19, this antibody-mediated phenomenon is detrimental (by suppressing a natural antiviral pathway, allowing uncontrolled virus replication) or beneficial (by reducing toxic effects of a potent antiviral response). That said, these results raise the possibility that therapy to block CD32B would partially restore interferon responses in people with severe disease. However, before considering therapeutic applications, the following steps should be taken. These results need to be confirmed in a larger group of patients, the process should be examined in other types of tissue in which the virus is found (rather than just in blood samples), and a fuller explanation is needed of the mechanisms that underlie these findings.
With several anti-SARS-CoV-2 vaccines currently approved, it will be useful to determine the antibody profile that vaccination elicits, and to compare it with the profile that develops during SARS-CoV-2 infection. Such a comparison would help to reveal the checks and balances used by the immune system to help keep us alive during severe infection.

 Read the paper: Global absence and targeting of protective immune states in severe COVID-19
Read the paper: Global absence and targeting of protective immune states in severe COVID-19
 Signs of self-sustained inflammatory circuits in severe COVID pneumonia
Signs of self-sustained inflammatory circuits in severe COVID pneumonia
 Interferon deficiency can lead to severe COVID
Interferon deficiency can lead to severe COVID