Abstract
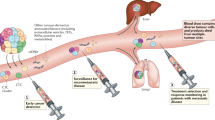
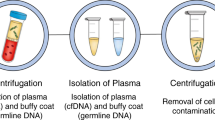
Liquid biopsy has been introduced as a new diagnostic concept predicated on the analysis of circulating tumour cells (CTCs) or circulating tumour-derived factors, in particular, cell-free tumour DNA (ctDNA). Highly sensitive liquid biopsy assays have been developed that can now be applied to detect and characterize minimal residual disease (MRD), which reflects the presence of tumour cells disseminated from the primary lesion to distant organs in patients who lack any clinical or radiological signs of metastasis or residual tumour cells left behind after local therapy that eventually lead to local recurrence. This application is the new frontier of liquid biopsy analyses, which are challenged by the very low concentrations of CTCs and ctDNA in blood samples. In this Review, we discuss the key technologies that can be used to detect and characterize CTCs in surveillance of MRD and provide a brief overview of similar roles of ctDNA analyses. We then focus on the current clinical data on the use of CTCs and ctDNA in the detection and monitoring of MRD and in obtaining information on therapeutic targets and resistance mechanisms relevant to the management of individual patients with cancer.
Key points
-
Minimal residual disease (MRD) can be defined as cancer persisting in a patient after treatment that cannot be detected with current medical imaging modalities and is, therefore, an occult stage of cancer progression.
-
Liquid biopsy approaches based on the detection of small numbers of circulating tumour cells (CTCs) or minute amounts of circulating cell-free tumour DNA (ctDNA) now enable MRD detection in patients with various malignancies.
-
CTC detection at primary diagnosis of cancer predicts an unfavourable prognosis and is, therefore, applicable to risk stratification strategies beyond the current approaches to tumour staging.
-
Monitoring of CTCs and ctDNA during post-surgical follow-up assessments can enable the detection of disease relapse many months earlier than is possible with current radiological imaging procedures.
-
Further characterization of CTCs and ctDNA can provide insights into the molecular evolution of MRD during tumour progression, with implications for therapy to delay or even prevent metastatic relapse.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Pan, H. et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 377, 1836–1846 (2017).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 36, 1631–1641 (2018).
Donaldson, J. & Park, B. H. Circulating tumor DNA: measurement and clinical utility. Annu. Rev. Med. 69, 223–234 (2018).
Heitzer, E., Ulz, P. & Geigl, J. B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 61, 112–123 (2015).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Alix-Panabieres, C., Mader, S. & Pantel, K. Epithelial-mesenchymal plasticity in circulating tumor cells. J. Mol. Med. 95, 133–142 (2017).
Ohnaga, T., Takei, Y., Nagata, T. & Shimada, Y. Highly efficient capture of cancer cells expressing EGFR by microfluidic methods based on antigen-antibody association. Sci. Rep. 8, 12005 (2018).
Thege, F. I. et al. Microfluidic immunocapture of circulating pancreatic cells using parallel EpCAM and MUC1 capture: characterization, optimization and downstream analysis. Lab. Chip 14, 1775–1784 (2014).
El-Heliebi, A. et al. In situ detection and quantification of AR-V7, AR-FL, PSA, and KRAS point mutations in circulating tumor cells. Clin. Chem. 64, 536–546 (2018).
Santana, S. M., Liu, H., Bander, N. H., Gleghorn, J. P. & Kirby, B. J. Immunocapture of prostate cancer cells by use of anti-PSMA antibodies in microdevices. Biomed. Microdevices 14, 401–407 (2012).
Hyun, K. A., Lee, T. Y., Lee, S. H. & Jung, H. I. Two-stage microfluidic chip for selective isolation of circulating tumor cells (CTCs). Biosens. Bioelectron. 67, 86–92 (2015).
Wu, L. L. et al. Chip-assisted single-cell biomarker profiling of heterogeneous circulating tumor cells using multifunctional nanospheres. Anal. Chem. 90, 10518–10526 (2018).
Paoletti, C. et al. Heterogeneous estrogen receptor expression in circulating tumor cells suggests diverse mechanisms of fulvestrant resistance. Mol. Oncol. 10, 1078–1085 (2016).
Coumans, F. A., van Dalum, G., Beck, M. & Terstappen, L. W. Filtration parameters influencing circulating tumor cell enrichment from whole blood. PLOS ONE 8, e61774 (2013).
Clawson, G. A. et al. Circulating tumor cells in melanoma patients. PLOS ONE 7, e41052 (2012).
Sun, N., Li, X., Wang, Z., Li, Y. & Pei, R. High-purity capture of CTCs based on micro-beads enhanced isolation by size of epithelial tumor cells (ISET) method. Biosens. Bioelectron. 102, 157–163 (2018).
Kim, T. H., Yoon, H. J., Stella, P. & Nagrath, S. Cascaded spiral microfluidic device for deterministic and high purity continuous separation of circulating tumor cells. Biomicrofluidics 8, 064117 (2014).
Huang, C., Smith, J. P., Saha, T. N., Rhim, A. D. & Kirby, B. J. Characterization of microfluidic shear-dependent epithelial cell adhesion molecule immunocapture and enrichment of pancreatic cancer cells from blood cells with dielectrophoresis. Biomicrofluidics 8, 044107 (2014).
Gascoyne, P. R. & Shim, S. Isolation of circulating tumor cells by dielectrophoresis. Cancers 6, 545–579 (2014).
Sarioglu, A. F. et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat. Methods 12, 685–691 (2015).
Gkountela, S. et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell 176, 98–112 (2019).
Heitzer, E. et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 73, 2965–2975 (2013).
Abonnenc, M. et al. Programmable interactions of functionalized single bioparticles in a dielectrophoresis-based microarray chip. Anal. Chem. 85, 8219–8224 (2013).
Peeters, D. J. et al. Semiautomated isolation and molecular characterisation of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting. Br. J. Cancer 108, 1358–1367 (2013).
Bhagwat, N. et al. An integrated flow cytometry-based platform for isolation and molecular characterization of circulating tumor single cells and clusters. Sci. Rep. 8, 5035 (2018).
Mastoraki, S. et al. ESR1 methylation: a liquid biopsy-based epigenetic assay for the follow-up of patients with metastatic breast cancer receiving endocrine treatment. Clin. Cancer Res. 24, 1500–1510 (2018).
Soler, A., Cayrefourcq, L., Mazel, M. & Alix-Panabieres, C. EpCAM-independent enrichment and detection of viable circulating tumor cells using the EPISPOT assay. Methods Mol. Biol. 1634, 263–276 (2017).
Ramirez, J. M. et al. Prognostic relevance of viable circulating tumor cells detected by EPISPOT in metastatic breast cancer patients. Clin. Chem. 60, 214–221 (2014).
Deneve, E. et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin. Chem. 59, 1384–1392 (2013).
Kuske, A. et al. Improved detection of circulating tumor cells in non-metastatic high-risk prostate cancer patients. Sci. Rep. 6, 39736 (2016).
Eyer, K. et al. Single-cell deep phenotyping of IgG-secreting cells for high-resolution immune monitoring. Nat. Biotechnol. 35, 977–982 (2017).
Gasch, C. et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin. Chem. 59, 252–260 (2013).
Carter, L. et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat. Med. 23, 114–119 (2017).
Lohr, J. G. et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat. Biotechnol. 32, 479–484 (2014).
Babayan, A. et al. Comparative study of whole genome amplification and next generation sequencing performance of single cancer cells. Oncotarget 8, 56066–56080 (2016).
Muller, C. et al. Hematogenous dissemination of glioblastoma multiforme. Sci. Transl Med. 6, 247ra101 (2014).
Yates, D. R. et al. Quantitative RT-PCR analysis of PSA and prostate-specific membrane antigen mRNA to detect circulating tumor cells improves recurrence-free survival nomogram prediction after radical prostatectomy. Prostate 72, 1382–1388 (2012).
Strati, A., Kasimir-Bauer, S., Markou, A., Parisi, C. & Lianidou, E. S. Comparison of three molecular assays for the detection and molecular characterization of circulating tumor cells in breast cancer. Breast Cancer Res. 15, R20 (2013).
Gorges, T. M. et al. Accession of tumor heterogeneity by multiplex transcriptome profiling of single circulating tumor cells. Clin. Chem. 62, 1504–1515 (2016).
Jordan, N. V. et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 537, 102–106 (2016).
Gasch, C. et al. Heterogeneity of miR-10b expression in circulating tumor cells. Sci. Rep. 5, 15980 (2015).
Sinkala, E. et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat. Commun. 8, 14622 (2017).
Franzen, B. et al. A fine-needle aspiration-based protein signature discriminates benign from malignant breast lesions. Mol. Oncol. 12, 1415–1428 (2018).
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Hodgkinson, C. L. et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 20, 897–903 (2014).
Zhang, L. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl Med. 5, 180ra48 (2013).
Yu, M. et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 345, 216–220 (2014).
Alix-Panabieres, C. et al. Molecular portrait of metastasis-competent circulating tumor cells in colon cancer reveals the crucial role of genes regulating energy metabolism and DNA repair. Clin. Chem. 63, 700–713 (2017).
Cayrefourcq, L. et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 75, 892–901 (2015).
Fischer, J. C. et al. Diagnostic leukapheresis enables reliable detection of circulating tumor cells of nonmetastatic cancer patients. Proc. Natl Acad. Sci. USA 110, 16580–16585 (2013).
Gorges, T. M. et al. Enumeration and molecular characterization of tumor cells in lung cancer patients using a novel in vivo device for capturing circulating tumor cells. Clin. Cancer Res. 22, 2197–2206 (2016).
Pantel, K. & Alix-Panabieres, C. Functional studies on viable circulating tumor cells. Clin. Chem. 62, 328–334 (2016).
Alix-Panabieres, C., Bartkowiak, K. & Pantel, K. Functional studies on circulating and disseminated tumor cells in carcinoma patients. Mol. Oncol. 10, 443–449 (2016).
Soler, A. et al. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci. Rep. 8, 15931 (2018).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2018).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC — challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Riethdorf, S. et al. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant “Geparquattro” trial. Clin. Cancer Res. 23, 5384–5393 (2017).
Bidard, F. C. et al. Circulating tumor cells in breast cancer patients treated by neoadjuvant chemotherapy: a meta-analysis. J. Natl Cancer Inst. 110, 560–567 (2018).
Janni, W. J. et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin. Cancer Res. 22, 2583–2593 (2016).
Rack, B. et al. Circulating tumor cells predict survival in early average-to-high risk breast cancer patients. J. Natl Cancer Inst. 106, dju066 (2014).
Yokobori, T. et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 73, 2059–2069 (2013).
Effenberger, K. E. et al. Improved risk stratification by circulating tumor cell counts in pancreatic cancer. Clin. Cancer Res. 24, 2844–2850 (2018).
Garcia-Murillas, I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl Med. 7, 302ra133 (2015).
Trapp, E. et al. Presence of circulating tumor cells in high-risk early breast cancer during follow-up and prognosis. J. Natl Cancer Inst. https://doi.org/10.1093/jnci/djy152 (2018).
Sparano, J. et al. Association of circulating tumor cells with late recurrence of estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 4, 1700–1706 (2018).
Goodman, C. R. et al. Association of circulating tumor cell status with benefit of radiotherapy and survival in early-stage breast cancer. JAMA Oncol. 4, e180163 (2018).
Bidard, F. C. et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol. 15, 406–414 (2014).
Heller, G. et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J. Clin. Oncol. 36, 572–580 (2018).
Ignatiadis, M. et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091–10093, BIG 1–12, Treat CTC): a randomized phase II trial. Ann. Oncol. 29, 1777–1783 (2018).
Georgoulias, V. et al. Trastuzumab decreases the incidence of clinical relapses in patients with early breast cancer presenting chemotherapy-resistant CK-19mRNA-positive circulating tumor cells: results of a randomized phase II study. Ann. Oncol. 23, 1744–1750 (2012).
van Dalum, G. et al. Importance of circulating tumor cells in newly diagnosed colorectal cancer. Int. J. Oncol. 46, 1361–1368 (2015).
Krebs, M. G. et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 29, 1556–1563 (2011).
Hou, J. M. et al. Clinical significance and molecular characteristics of circulating tumor cells and circulating tumor microemboli in patients with small-cell lung cancer. J. Clin. Oncol. 30, 525–532 (2012).
Aggarwal, C. et al. Circulating tumor cells as a predictive biomarker in patients with small cell lung cancer undergoing chemotherapy. Lung Cancer 112, 118–125 (2017).
Lindsay, C. R. et al. A prospective examination of circulating tumor cell profiles in non-small-cell lung cancer molecular subgroups. Ann. Oncol. 28, 1523–1531 (2017).
Scher, H. I. et al. Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur. Urol. 71, 874–882 (2017).
Meng, S. et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 10, 8152–8162 (2004).
Chen, Y. H. et al. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. NPJ Breast Cancer 3, 24 (2017).
Olsson, E. et al. Serial monitoring of circulating tumor DNA in patients with primary breast cancer for detection of occult metastatic disease. EMBO Mol. Med. 7, 1034–1047 (2015).
Riva, F. et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin. Chem. 63, 691–699 (2017).
Tie, J. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl Med. 8, 346ra92 (2016).
Ng, S. B. et al. Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery. Sci. Rep. 7, 40737 (2017).
Scholer, L. V. et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 23, 5437–5445 (2017).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Kuang, Y. et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin. Cancer Res. 15, 2630–2636 (2009).
Nakamura, T. et al. A noninvasive system for monitoring resistance to epidermal growth factor receptor tyrosine kinase inhibitors with plasma DNA. J. Thorac. Oncol. 6, 1639–1648 (2011).
Taniguchi, K. et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin. Cancer Res. 17, 7808–7815 (2011).
Douillard, J. Y. et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br. J. Cancer 110, 55–62 (2014).
QIAGEN. therascreen® EGFR RGQ PCR kit instructions for use (handbook). FDA https://www.accessdata.fda.gov/cdrh_docs/pdf12/P120022c.pdf (2013).
Maheswaran, S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 359, 366–377 (2008).
Mostert, B. et al. KRAS and BRAF mutation status in circulating colorectal tumor cells and their correlation with primary and metastatic tumor tissue. Int. J. Cancer 133, 130–141 (2013).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl Med. 6, 224ra24 (2014).
Siravegna, G. et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 21, 827 (2015).
Jiang, Y., Palma, J. F., Agus, D. B., Wang, Y. & Gross, M. E. Detection of androgen receptor mutations in circulating tumor cells in castration-resistant prostate cancer. Clin. Chem. 56, 1492–1495 (2010).
Wyatt, A. W. et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2, 1598–1606 (2016).
Meric-Bernstam, F. et al. Advances in HER2-targeted therapy: novel agents and opportunities beyond breast and gastric cancer. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-18-2275 (2019).
Schneck, H. et al. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 7, 976–986 (2013).
Neves, R. P. et al. Genomic high-resolution profiling of single CKpos/CD45neg flow-sorting purified circulating tumor cells from patients with metastatic breast cancer. Clin. Chem. 60, 1290–1297 (2014).
Pestrin, M. et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol. Oncol. 9, 749–757 (2015).
Polzer, B. et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 6, 1371–1386 (2014).
Gasch, C. et al. Frequent detection of PIK3CA mutations in single circulating tumor cells of patients suffering from HER2-negative metastatic breast cancer. Mol. Oncol. 10, 1330–1343 (2016).
Schiavon, G. et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci. Transl Med. 7, 313ra182 (2015).
O’Leary, B. et al. Early circulating tumor DNA dynamics and clonal selection with palbociclib and fulvestrant for breast cancer. Nat. Commun. 9, 896 (2018).
Robinson, D. R. et al. Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat. Genet. 45, 1446–1451 (2013).
Toy, W. et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 45, 1439–1445 (2013).
Shaw, J. A. et al. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin. Cancer Res. 23, 88–96 (2017).
Guttery, D. S. et al. Noninvasive detection of activating estrogen receptor 1 (ESR1) mutations in estrogen receptor-positive metastatic breast cancer. Clin. Chem. 61, 974–982 (2015).
Chu, D. et al. ESR1 mutations in circulating plasma tumor DNA from metastatic breast cancer patients. Clin. Cancer Res. 22, 993–999 (2016).
O’Leary, B. et al. The genetic landscape and clonal evolution of breast cancer resistance to palbociclib plus fulvestrant in the PALOMA-3 trial. Cancer Discov. 8, 1390–1403 (2018).
Villanueva, J., Vultur, A. & Herlyn, M. Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 71, 7137–7140 (2011).
Long, G. V. et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 17, 1743–1754 (2016).
Sakaizawa, K. et al. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br. J. Cancer 106, 939–946 (2012).
Long, G. V. et al. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N. Engl. J. Med. 377, 1813–1823 (2017).
Pailler, E. et al. Method for semi-automated microscopy of filtration-enriched circulating tumor cells. BMC Cancer 16, 477 (2016).
Pailler, E. et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J. Clin. Oncol. 31, 2273–2281 (2013).
Pailler, E. et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann. Oncol. 26, 1408–1415 (2015).
Attard, G. et al. Characterization of ERG, AR and PTEN gene status in circulating tumor cells from patients with castration-resistant prostate cancer. Cancer Res. 69, 2912–2918 (2009).
Danila, D. C. et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur. Urol. 60, 897–904 (2011).
Antonarakis, E. S. et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 371, 1028–1038 (2014).
Steinestel, J. et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget https://doi.org/10.18632/oncotarget.3925 (2015).
Thadani-Mulero, M. et al. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 74, 2270–2282 (2014).
Antonarakis, E. S. et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 1, 582–591 (2015).
Nakazawa, M. et al. Serial blood-based analysis of AR-V7 in men with advanced prostate cancer. Ann. Oncol. 26, 1859–1865 (2015).
Onstenk, W. et al. Efficacy of cabazitaxel in castration-resistant prostate cancer is independent of the presence of AR-V7 in circulating tumor cells. Eur. Urol. 68, 939–945 (2015).
Scher, H. I. et al. Assessment of the validity of nuclear-localized androgen receptor splice variant 7 in circulating tumor cells as a predictive biomarker for castration-resistant prostate cancer. JAMA Oncol. 4, 1179–1186 (2018).
Miyamoto, D. T. et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 349, 1351–1356 (2015).
Maurer, T. et al. PSMA theranostics using PET and subsequent radioguided surgery in recurrent prostate cancer. Clin. Genitourin. Cancer 14, e549–e552 (2016).
Ristau, B. T., O’Keefe, D. S. & Bacich, D. J. The prostate-specific membrane antigen: lessons and current clinical implications from 20 years of research. Urol. Oncol. 32, 272–279 (2014).
Ahmadzadehfar, H. et al. Predictors of overall survival in metastatic castration-resistant prostate cancer patients receiving [(177)Lu]Lu-PSMA-617 radioligand therapy. Oncotarget 8, 103108–103116 (2017).
Babayan, A. et al. Heterogeneity of estrogen receptor expression in circulating tumor cells from metastatic breast cancer patients. PLOS ONE 8, e75038 (2013).
Paoletti, C. et al. Development of circulating tumor cell-endocrine therapy index in patients with hormone receptor-positive breast cancer. Clin. Cancer Res. 21, 2487–2498 (2015).
Fehm, T. et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res. Treat. 124, 403–412 (2010).
Wang, C. H., Chang, C. J., Yeh, K. Y., Chang, P. H. & Huang, J. S. The prognostic value of HER2-positive circulating tumor cells in breast cancer patients: a systematic review and meta-analysis. Clin. Breast Cancer 17, 341–349 (2017).
Philips, G. K. & Atkins, M. Therapeutic uses of anti-PD-1 and anti-PD-L1 antibodies. Int. Immunol. 27, 39–46 (2015).
Mittendorf, E. A. et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2, 361–370 (2014).
Mazel, M. et al. Frequent expression of PD-L1 on circulating breast cancer cells. Mol. Oncol. 9, 1773–1782 (2015).
Nicolazzo, C. et al. Monitoring PD-L1 positive circulating tumor cells in non-small cell lung cancer patients treated with the PD-1 inhibitor nivolumab. Sci. Rep. 6, 31726 (2016).
Strati, A. et al. Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 28, 1923–1933 (2017).
Cabel, L. et al. Clinical potential of circulating tumour DNA in patients receiving anticancer immunotherapy. Nat. Rev. Clin. Oncol. 15, 639–650 (2018).
Mohme, M., Riethdorf, S. & Pantel, K. Circulating and disseminated tumour cells — mechanisms of immune surveillance and escape. Nat. Rev. Clin. Oncol. 14, 155–167 (2017).
Pantel, K. & Hayes, D. F. Disseminated breast tumour cells: biological and clinical meaning. Nat. Rev. Clin. Oncol. 15, 129–131 (2018).
Ulz, P. et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 48, 1273–1278 (2016).
Guo, N. et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 6, 33519 (2016).
Alix-Panabieres, C. & Pantel, K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 6, 479–491 (2016).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Mani, S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133, 704–715 (2008).
Fernandez-Cuesta, L. et al. Identification of circulating tumor DNA for the early detection of small-cell lung cancer. EBioMedicine 10, 117–123 (2016).
Krimmel, J. D. et al. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc. Natl Acad. Sci. USA 113, 6005–6010 (2016).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Kato, S., Lippman, S. M., Flaherty, K. T. & Kurzrock, R. The conundrum of genetic “drivers” in benign conditions. J. Natl Cancer Inst. 108, djw036 (2016).
Pollock, P. M. et al. High frequency of BRAF mutations in nevi. Nat. Genet. 33, 19–20 (2003).
Bardelli, A. & Pantel, K. Liquid biopsies, what we do not know (yet). Cancer Cell 31, 172–179 (2017).
Gao, X. L., Zhang, M., Tang, Y. L. & Liang, X. H. Cancer cell dormancy: mechanisms and implications of cancer recurrence and metastasis. Onco Targets Ther. 10, 5219–5228 (2017).
Acknowledgements
The authors thank N. Reimers for her assistance in designing figure 4. The authors received support from the German Cancer Aid Fund (Deutsche Krebshilfe); DFG (Deutsche Forschungsgemeinschaft); the French National Institute of Cancer (INCa); CANCER-ID, an Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115749, resources of which are composed of financial contribution from the European Union’s Seventh Framework Program (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies’ in-kind contributions; and the European Liquid Biopsies Academy (ELBA) Innovative Training Networks (ITN) Horizon 2020 project H2020-MSCA-ITN-2017 (Towards widespread clinical application of blood-based diagnostic tools).
Competing interests
K.P. and C.A.-P. have ongoing patent applications related to circulating tumour cells. K.P. has received honoraria from Agena, Novartis, Roche and Sanofi and research funding from European Federation of Pharmaceutical Industries and Associations (EFPIA) partners (Angle, Menarini and Servier) of the CANCER-ID programme of the European Union–EFPIA Innovative Medicines Initiative. C.A.-P. has received honoraria from Janssen.
Reviewer information
Nature Reviews Clinical Oncology thanks J.-Y. Pierga and other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Both authors made substantial contributions to all stages of the preparation of this manuscript.
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
BloodPAC: https://www.bloodpac.org/
CANCER-ID: https://www.cancer-id.eu/
ClinicalTrials.gov database: https://clinicaltrials.gov/
Supplementary information
Rights and permissions
About this article
Cite this article
Pantel, K., Alix-Panabières, C. Liquid biopsy and minimal residual disease — latest advances and implications for cure. Nat Rev Clin Oncol 16, 409–424 (2019). https://doi.org/10.1038/s41571-019-0187-3
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41571-019-0187-3
This article is cited by
-
c-MET-positive circulating tumor cells and cell-free DNA as independent prognostic factors in hormone receptor-positive/HER2-negative metastatic breast cancer
Breast Cancer Research (2024)
-
A multiparameter liquid biopsy approach allows to track melanoma dynamics and identify early treatment resistance
npj Precision Oncology (2024)
-
Management of patients with advanced-stage HER2-positive breast cancer: current evidence and future perspectives
Nature Reviews Clinical Oncology (2024)
-
Changes in cell-free DNA after short-term palbociclib and fulvestrant treatment for advanced or metastatic hormone receptor-positive and human epidermal growth factor 2-negative breast cancer
Breast Cancer Research and Treatment (2024)
-
Clinical applications of circulating tumor cells in patients with solid tumors
Clinical & Experimental Metastasis (2024)