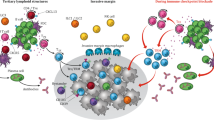
Abstract
Treatment with immune checkpoint blockade (ICB) has revolutionized cancer therapy. Until now, predictive biomarkers1,2,3,4,5,6,7,8,9,10 and strategies to augment clinical response have largely focused on the T cell compartment. However, other immune subsets may also contribute to anti-tumour immunity11,12,13,14,15, although these have been less well-studied in ICB treatment16. A previously conducted neoadjuvant ICB trial in patients with melanoma showed via targeted expression profiling17 that B cell signatures were enriched in the tumours of patients who respond to treatment versus non-responding patients. To build on this, here we performed bulk RNA sequencing and found that B cell markers were the most differentially expressed genes in the tumours of responders versus non-responders. Our findings were corroborated using a computational method (MCP-counter18) to estimate the immune and stromal composition in this and two other ICB-treated cohorts (patients with melanoma and renal cell carcinoma). Histological evaluation highlighted the localization of B cells within tertiary lymphoid structures. We assessed the potential functional contributions of B cells via bulk and single-cell RNA sequencing, which demonstrate clonal expansion and unique functional states of B cells in responders. Mass cytometry showed that switched memory B cells were enriched in the tumours of responders. Together, these data provide insights into the potential role of B cells and tertiary lymphoid structures in the response to ICB treatment, with implications for the development of biomarkers and therapeutic targets.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The additional datasets generated during and/or analysed during the current study for Clinical Trial NCT02519322 are now available in the European Genome-phenome Archive repository (EGAS00001003178). Other datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Code availability
The authors declare that the code for reproducibility of data are publicly available or will be available upon request.
References
Chen, P. L. et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 6, 827–837 (2016).
Taube, J. M. et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin. Cancer Res. 20, 5064–5074 (2014).
Cottrell, T. R. & Taube, J. M. PD-L1 and emerging biomarkers in immune checkpoint blockade therapy. Cancer J. 24, 41–46 (2018).
Yarchoan, M., Hopkins, A. & Jaffee, E. M. Tumor mutational burden and response rate to PD-1 inhibition. N. Engl. J. Med. 377, 2500–2501 (2017).
Ayers, M. et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Invest. 127, 2930–2940 (2017).
Subudhi, S. K. et al. Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc. Natl Acad. Sci. USA 113, 11919–11924 (2016).
Jacquelot, N. et al. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat. Commun. 8, 592 (2017).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018).
Fridman, W. H., Zitvogel, L., Sautès-Fridman, C. & Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14, 717–734 (2017).
Colbeck, E. J., Ager, A., Gallimore, A. & Jones, G. W. Tertiary lymphoid structures in cancer: drivers of antitumor immunity, immunosuppression, or bystander sentinels in disease? Front. Immunol. 8, 1830 (2017).
Dieu-Nosjean, M. C., Goc, J., Giraldo, N. A., Sautès-Fridman, C. & Fridman, W. H. Tertiary lymphoid structures in cancer and beyond. Trends Immunol. 35, 571–580 (2014).
Sarvaria, A., Madrigal, J. A. & Saudemont, A. B cell regulation in cancer and anti-tumor immunity. Cell. Mol. Immunol. 14, 662–674 (2017).
Tsou, P., Katayama, H., Ostrin, E. J. & Hanash, S. M. The emerging role of B cells in tumor immunity. Cancer Res. 76, 5597–5601 (2016).
Sautès-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325 (2019).
Amaria, R. N. et al. Neoadjuvant immune checkpoint blockade in high-risk resectable melanoma. Nat. Med. 24, 1649–1654 (2018).
Becht, E. et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 17, 218 (2016).
Yuen, G. J., Demissie, E. & Pillai, S. B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2, 747–757 (2016).
Chiaruttini, G. et al. B cells and the humoral response in melanoma: the overlooked players of the tumor microenvironment. OncoImmunology 6, e1294296 (2017).
Erdag, G. et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res. 72, 1070–1080 (2012).
Iglesia, M. D. et al. Genomic analysis of immune cell infiltrates across 11 tumor types. J. Natl. Cancer Inst. 108, (2016).
Ladányi, A. et al. Prognostic impact of B-cell density in cutaneous melanoma. Cancer Immunol. Immunother. 60, 1729–1738 (2011).
Garg, K. et al. Tumor-associated B cells in cutaneous primary melanoma and improved clinical outcome. Hum. Pathol. 54, 157–164 (2016).
Ladányi, A. et al. Density of DC-LAMP+ mature dendritic cells in combination with activated T lymphocytes infiltrating primary cutaneous melanoma is a strong independent prognostic factor. Cancer Immunol. Immunother. 56, 1459–1469 (2007).
Martinet, L. et al. High endothelial venules (HEVs) in human melanoma lesions: Major gateways for tumor-infiltrating lymphocytes. OncoImmunology 1, 829–839 (2012).
Avram, G. et al. The density and type of MECA-79-positive high endothelial venules correlate with lymphocytic infiltration and tumour regression in primary cutaneous melanoma. Histopathology 63, 852–861 (2013).
Messina, J. L. et al. 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci. Rep. 2, 765 (2012).
Goc, J. et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 74, 705–715 (2014).
Posch, F. et al. Maturation of tertiary lymphoid structures and recurrence of stage II and III colorectal cancer. OncoImmunology 7, e1378844 (2017).
Siliņa, K. et al. Germinal centers determine the prognostic relevance of tertiary lymphoid structures and are impaired by corticosteroids in lung squamous cell carcinoma. Cancer Res. 78, 1308–1320 (2018).
Cipponi, A. et al. Neogenesis of lymphoid structures and antibody responses occur in human melanoma metastases. Cancer Res. 72, 3997–4007 (2012).
Selitsky, S. R. et al. Prognostic value of B cells in cutaneous melanoma. Genome Med. 11, 36 (2019).
Griss, J. et al. B cells sustain inflammation and predict response to immune checkpoint blockade in human melanoma. Nat. Commun. 10, 4186 (2019).
Blank, C. U. et al. Neoadjuvant versus adjuvant ipilimumab plus nivolumab in macroscopic stage III melanoma. Nat. Med. 24, 1655–1661 (2018).
Cabrita, R. L. et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature https://doi.org/10.1038/s41586-019-1914-8 (2020).
Petitprez, F. et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature https://doi.org/10.1038/s41586-019-1906-8 (2020).
Amaria, R. N. et al. Neoadjuvant plus adjuvant dabrafenib and trametinib versus standard of care in patients with high-risk, surgically resectable melanoma: a single-centre, open-label, randomised, phase 2 trial. Lancet Oncol. 19, 181–193 (2018).
Reddy, S. M. et al. Poor response to neoadjuvant chemotherapy correlates with mast cell infiltration in inflammatory breast cancer. Cancer Immunol. Res. 7, 1025–1035 (2019).
Song, I. H. et al. Predictive value of tertiary lymphoid structures assessed by high endothelial venule counts in the neoadjuvant setting of triple-negative breast cancer. Cancer Res. Treat. 49, 399–407 (2017).
Germain, C. et al. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am. J. Respir. Crit. Care Med. 189, 832–844 (2014).
Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell 161, 1681–1696 (2015).
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49 (2013).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013 (2018).
FastQC. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Love, M. I., Anders, S., Kim, V. & Huber, W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000 Res. 4, 1070 (2015).
Wu, G., Dawson, E., Duong, A., Haw, R. & Stein, L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000 Res. 3, 146 (2014).
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Liu, J. et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416 (2018).
Benjamini, Y., Drai, D., Elmer, G., Kafkafi, N. & Golani, I. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 (2001).
Pimenta, E. M. & Barnes, B. J. Role of tertiary lymphoid structures (TLS) in anti-tumor immunity: potential tumor-induced cytokines/chemokines that regulate TLS formation in epithelial-derived cancers. Cancers (Basel) 6, 969–997 (2014).
Hiraoka, N. et al. Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br. J. Cancer 112, 1782–1790 (2015).
Ruddle, N. H. Lymphatic vessels and tertiary lymphoid organs. J. Clin. Invest. 124, 953–959 (2014).
Hu, X. et al. Landscape of B cell immunity and related immune evasion in human cancers. Nat. Genet. 51, 560–567 (2019).
Nazarov, V. I. et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinformatics 16, 175 (2015).
Villani, A. C. et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356, eaah4573 (2017).
Acknowledgements
The correlative research was funded by philanthropic support from the MD Anderson Melanoma Moon Shot Program, the Parker Institute for Cancer Immunotherapy, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the AIM at Melanoma Foundation. It was supported by The Immunotherapy Platform and the Core grant CA016672 (SMF) to support the Sequencing and Microarray Facility at UT-MDACC as well as the the Office of the Assistant Secretary for Defense for Health Affairs grant (W81XWH-16-1-0120 and W81XWH-16-1-0121). S.M.R. received support from National Institutes of Health T32 CA 009666 and Cancer Prevention Research Institute of Texas RR190020. B.A.H. received support from National Institutes of Health T32 CA 009599 and the MD Anderson Cancer Center support grant P30 CA016672. A.P.C. is supported by the CPRIT Research Training Program (RP170067) and the United States Department of State’s Bureau of Educational and Cultural Affairs. E.Z.K. is supported by National Institutes of Health grant T32 CA009599. F.G.K. is supported with funding from the Odyssey Program at the MD Anderson Cancer Center with support from the Theodore N. Law Endowment for Scientific Achievement. Research in the Kalluri laboratory is supported by the Cancer Prevention and Research Institute of Texas and National Cancer Institute grants CA213233, P01CA117969, and CA195733. K.M.M. and M.A.Z. are supported by CPRIT RP190507. The High Resolution Electron Microscopy Facility at UT-MDASS is supported by CCSG grant NIGH P30CA016672. We thank to O. Contrares for technical support on the multiplex immunofluorescence and for M. Andrews for technical support on RNA-seq library preparation and K. McAndrews, D. P. Dowlatshahi, L. Snowden, J. Leveille and S. Yang for support with analyses of exosomes.
Author information
Authors and Affiliations
Contributions
B.A.H., S.M.R., J.G., S.Z. and R.B. contributed equally. Jointly supervising authors include P.S., K.R., M.T.T., L.W. and J.A.W. Corresponding authors are B.A.H. and J.A.W. J.A.W. designed the study. J.E.G., R.N.A., P.H., M.A.D., H.A.T., S.P., A.L., S. Woodman, C.U.B., T.N.S., P.S. and J.A.W. collected samples for correlative analyses. B.A.H., S.M.R., J.G., S.Z., R.B., R.T., K.Y., M.S.-F., J.B., G.H., V.G., Y.X., H.Z., A.P.C., W.L., V.S.L., F.G.K., A.L., R.A., S. Woodman, E.Z.K., P.-O.G., A.R., C.N.S., E.M.B., L.E.H., A.J.L., R.Z., C.W.H., D.A.L., S.O., M.B., S. Warren, D.R., O.K., E.A.R., D.P., C.U.B., T.N.S., L.H.B., M.A.Z., K.M.M., R.K., J.A., F.P., W.H.F., C.S.-F. and N.H. analysed and interpreted data. Specifically, K.Y., M.S.F. and N.H. assisted with single-cell RNA-seq analyses. S.O., M.B. and S. Warren assisted with Nanostring Digital Spatial Profiling analyses. R.B. and K.R. assisted with CyTOF. S.Z., R.T., L.W., M.A.Z. and K.M.M. assisted with BCR analyses. J.B., S.O., M.B., S. Warren, C.H., D.L. and M.T.T. assisted with singlet and multifocal immunohistochemistry. S.Z., L.W., J.E.G. and A.J.L. assisted with analyses of the TCGA patient cohorts. S.Z., R.T. and L.W. assisted with all bioinformatics and statistical analyses. R.T. performed pathway analyses. V.S.L., F.G.K. and R.K. assisted with analyses of exosomes. D.R., O.K., E.A.R., D.P., C.U.B. and T.N.S. provided assistance with analyses of patients from the OpACIN-neo trial. J.G., J.B. and P.S. assisted with analyses of the RCC patient cohort. All authors developed and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.A.W. is an inventor on a US patent application (PCT/US17/53.717) submitted by the University of Texas MD Anderson Cancer Center that covers methods to enhance immune checkpoint blockade responses by modulating the microbiome. J.A.W. reports compensation for speaker’s bureau and honoraria from Imedex, Dava Oncology, Omniprex, Illumina, Gilead, PeerView, Physician Education Resource, MedImmune and Bristol-Myers Squibb. J.A.W. serves as a consultant/advisory board member for Roche/Genentech, Novartis, AstraZeneca, GlaxoSmithKline, Bristol-Myers Squibb, Merck, Biothera Pharmaceuticals and Microbiome DX. J.A.W. also receives research support from GlaxoSmithKline, Roche/Genentech, Bristol-Myers Squibb, and Novartis. J.A.W., S.M.R. and B.A.H. are co-inventors on an unpublished patent application related to methods of targeting B cells to enhance response to immune checkpoint blockade. M.T.T. reports advisory board participation and speaker paid honorarium from Nanostring and Myriad Genetics. M.A.D. serves as a paid consultant for BMS, Novartis, and Roche/Genentech. M.A.D. also reports to be a principal investigator of a research grant from Roche/Genentech and an unpaid consultant to Nanostring. C.U.B. reports an advisory role in BMS, MSD, Roche, Novartis, GSK, AZ, Pfizer, GenMab and Pierre Fabre. C.U.B. receives research funding from BMS, Novartis and Nanostring. C.U.B. reports stock ownership from Uniti Cars and Neon Therapeutics. N.H. is a founder, stockholder and SAB member of Neon Therapeutics. W.H.F. serves as a consultant for AstraZeneca, Ipsen, Adaptimmune, OxfordBiotherapeutics, and Catalym. W.H.F. reports participation in data transparency committee for Servier and data management committee for Novartis. O.K. receives grant support from BMS. J.E.G. is a contributor of UpToDate-melanoma staging and prognosis. J.E.G. reports to be an unpaid member of Melanoma Research Foundation and Melanoma Research Alliance. J.E.G. reports to be on advisory board of Merck. R.K. reports to be scientific founder, stockholder and consultant of Codiak Biosciences. A.J.L. reports consultancies and research support from BMS, Genentech/Roche and MedImmune/Astra-Zeneca. P.S. reports a patent licensed to Jounce Therapeutics. P.S. serves as a consultant for Constellation, Jounce Therapeutics, Neon, BioAtla, Pieris, Oncolytics, Forty-Seven, Polaris, Apricity, Marker, Codiak, ImaginAb, Hummingbird, Dragonfly, Lytix and Bristol-Myers Squibb (BMS). P.S. has ownership interests in Jounce Therapeutics, Neon, Constellation, Oncolytics, BioAtla, Forty-Seven, Apricity, Polaris, Marker, Codiak, ImaginAb, Hummingbird, Dragonfly and Lytix. L.H.B. reports to be on StemImmune/Calidi Scientific and Medical Advisory, Scientific Advisory Board of BoardNextCure, Replimmune, Western Oncolytics, Torque Therapeutics Khloris, Pyxis, Cytomix. L.H.B. reports to be Chair of Food and Drug Administration Cellular, Tissues and Gene Therapies Advisory Committee. R.Z. reports a patent application related to work on GITR, PD1 and CTLA4. R.Z. is a consultant for Leap Therapeutics. P.H. is on advisory board for Dragonfly, GlaxoSmithKline, Immatics and Sanofi. S.W., S.O. and M.B. are employees and stockholders of NanoString Technologies. All other authors report no competing interests directly relevant to this work.
Additional information
Peer review information Nature thanks James Mulé, Caroline Robert and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 MCP-counter results in patients with melanoma and RCC treated with pre-surgical ICB or targeted therapy.
a, Supervised clustering by response of MCP-counter scores in on-treatment samples from a cohort of high-risk patients with resectable melanoma treated with neoadjuvant ICB, with responders defined as achieving a complete or partial response by RECIST 1.1 (n = 11 NR and 9 R). b, Analysis shown by unsupervised hierarchical clustering of baseline (n = 11 NR and 10 R) and on-treatment samples (n = 11 NR and 9R) from the neoadjuvant melanoma cohort. Unique clusters identified are indicated by shaded boxes on top row. c, Unsupervised hierarchical analysis shown for metastatic RCC patients (same cohort as Fig. 1d; n = 11 PD and 17 PR). Response (PR, partial response) or non-response (PD, progressive disease) as measured by RECIST 1.1. Unique clusters identified are indicated by shaded boxes on top row. d, Supervised clustering by response of MCP-counter scores from OpACIN-neo clinical trial (NCT02437279) of neoadjuvant versus adjuvant ICB in high-risk resectable melanoma (n = 6 NR and 12 R). Responders were defined as patients who did not have a relapse. e, Supervised clustering by response of MCP-counter scores in combined pre-treatment and on-treatment biopsies from a cohort of high-risk resectable melanoma patients treated with neoadjuvant targeted therapy (dabrafenib and trametinib) as part of NCT02231775 (n = 7 patients for baseline and n = 8 patients for on-treatment samples) with responder defined as achieving a complete or partial response by RECIST 1.1 and non-responder defined as having stable or progressive disease. Pathological response is defined by the presence or absence of viable tumour at time of surgical resection. P values were made using two-sided Mann–Whitney U-test.
Extended Data Fig. 2 Representation of MCP-counter scores for all patient cohorts and analyses of peripheral blood exosomes.
a–c, Box plot representation of heat maps for patients with: high-risk resectable melanoma treated with neoadjuvant ICB (n = 11 NR and 10 R for baseline and n = 11 NR and 9 R on treatment) as presented in Fig. 1c and Extended Data Fig. 1a, b (a); metastatic RCC treated with pre-surgical ICB as presented in Fig. 1d and Extended Data Fig. 1c (n = 11 PD and 17 PR) (b); and high-risk resectable melanoma treated with ICB as part of OpACIN-neo trial as presented in Extended Data Fig. 1d (n = 6 NR and 12 R) (c). For a–c, medians with interquartile range are shown. P values were determined by two-sided Mann–Whitney U-test. d, Schematic for exosomal analyses of serum samples from patients with melanoma on neoadjuvant ICB trial. e, Representative transmission electron micrographs showing Dynabead with exosomes present after immunocapture. f, Nanoimager-captured images of the beads coated with CD63+ exosomes as compared with isotype control. g, h, Exosomal concentration (g) and mean exosomal size (h) for serum samples for responders and non-responders at the time point indicated. i, Ratio of mean fluorescent intensity (MFI) of beads stained with anti-CD63 as compared to isotype control. j, Ratio of mean fluorescent intensity of beads stained with anti-CD9, -CD20, -CD27 and -GPC1 antibodies as compared to isotype control (or secondary antibody only for GPC1). For e–j, bars indicate median values and individual data points representing 8 R and 5 NR (unless indicated in the Methods) in addition to interquartile ranges. P values were determined using two-sided Mann–Whitney U-test.
Extended Data Fig. 3 Transcriptional analysis of tumour specimens from patients with metastatic RCC treated with pre-surgical ICB.
a, Supervised hierarchical clustering by response of RCC tumour specimens at baseline of most DEGs by microarray analysis, with response defined as having a partial response by RECIST 1.1 and non-response as having progressive disease (n = 11 PD and 17 PR). Fold change and P values are calculated by the limma package as described in the Methods. A cut-off of gene expression fold change of ≥ 2 or ≤ 0.5 and a FDR q ≤ 0.05 was applied to select DEGs. b, Volcano plot depiction of DEGs by response from same cohort.
Extended Data Fig. 4 Immune infiltrate is prognostic of improved disease-specific survival in TCGA cutaneous melanoma cohort but not the clear-cell RCC cohort.
a, Unsupervised hierarchical analysis of TCGA SKCM RNA-seq data using MCP-counter scores identifies three MICs with differential presence of individual cell types as indicated. Numbers of patients in each class is shown on top of the plot. P value determined by two-sided Kruskal–Wallis rank-sum test and q value calculated by FDR. b, Kaplan–Meier estimates of overall survival of MIC groups. c, Kaplan–Meier estimates of overall survival by B cell lineage scores shown by high and low groups dichotomized by median values. Overall survival was defined as the time interval from date of accession for each sample to date of death or censoring from any cause (Methods). d, Unsupervised hierarchical analysis of TCGA KIRC RNA-seq data using MCP-counter scores identifies three immune classes with differential presence of individual cell types as indicated. Numbers of patients in each class are shown at top of plot. P values determined by two-sided Kruskal–Wallis rank-sum test q value calculated by FDR. e, Kaplan–Meier estimates of overall survival probability of immune class groups. f, Kaplan–Meier estimates of overall survival probability by B cell lineage scores shown by high and low groups dichotomized by median values. For both, overall survival was defined as the time interval from date of accession for each sample to date of death or censoring from any cause. For b, c, e, f, patient numbers are included in the table below the graph and P values were calculated by log-rank test.
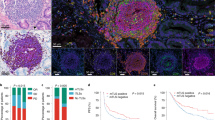
Extended Data Fig. 5 TLSs found in nodal and non-nodal metastases are consistent with mature secondary follicular-like TLSs with modest correlation with gene expression data.
a, Representative TLSs in tumours from patients with melanoma treated with neoadjuvant ICB demonstrating maturation status as indicated by the presence of follicular dendritic cells (CD21) and germinal centre B cells (CD23). We also include multiplex immunohistochemistry for SYTO13, MECA79, CD20 and CD4 (with magnified view of individual TLSs indicated by white box on the right). Circles denote defined TLSs based on multiplex immunohistochemistry. Black line approximates tumour border. b, Representative TLSs from non-lymph node metastases on additional patients with metastatic melanoma indicated by H&E staining, as well as singlet staining for CD20, CD21 and CD23. Black line on H&E image denotes tumour border. c, Comparison of baseline and on-treatment gene expression with MCP-counter analyses for B cell lineage as well as MS4A1 expression by RNA-seq for patients with high-risk resectable melanoma treated with ICB as part of clinical trial (n = 11 NR and 10 R for baseline and n = 11 NR and 9 R for on-treatment). Response and treatment arm as indicated. Medians with interquartile range are shown. P values were determined by two-sided Mann–Whitney U-test. d, Linear regression modelling of MCP-counter values for B cell lineage with regards to CD20 counts (n = 10 NR and 7 R) and TLS density (n = 10 NR and 6 R) as indicated. e, Linear regression modelling of MS4A1 gene expression with regards to CD20 counts (n = 10 NR and 7 R) and TLS density (n = 10 NR and 6 R) as indicated. These represent on-treatment time points. For d, e, r, values calculated by linear regression and P values for non-zero slope as calculated by Prism v.8.0.0.
Extended Data Fig. 6 TLSs are associated with response in RCC similar to those observed in melanoma.
a, Multiplex immunohistochemistry images from three additional patients with advanced melanoma treated with neoadjuvant ICB. Staining as indicated and similar to Fig. 2. b, Quantification of CD20 cells by singlet immunohistochemistry and association with response to neoadjuvant ICB in metastatic RCC, with responders defined as having partial response and non-responders as having progressive disease by RECIST 1.1 (n = 10 PD and 8 PR at baseline and n = 5 PD and 11 PR on treatment). c, d, Density of TLSs (n = 10 PD and 9 PR at baseline and n = 5 PD and 9 PR on treatment) (c) and ratio of tumour area occupied by TLSs (n = 10 PD and 7 PR at baseline and n = 5 PD and 11 PR on treatment) (d) and correlation by treatment response. Bars indicate median values and interquartile ranges are shown. P values were determined by two-sided Mann–Whitney U-test. e–g, Representative image of CD20 staining in responder with TLSs, associated H&E slide, singlet stains and characterization by multiplex immunofluorescence of TLSs. h, Multiplex immunohistochemistry images from three additional patients with RCC treated with pre-surgical ICB. Staining as indicated and similar to g.
Extended Data Fig. 7 TLSs are associated with markers of T cell activation and response and B cell proliferation.
NanoString GeoMx Digital Spatial Profiling was used to perform high-plex proteomic analysis with spatial resolution. a, Example of selection of ROIs (200 μm × 200 μm) from representative patients with melanoma treated with neoadjuvant ICB including ROI containing TLSs and ROIs outside the context of a TLS (non-TLS). ROI selection was completed using H&E staining and confirmed with immunofluorescence as shown using S100B and PMEL, SYTO13, CD3 and CD20. Masking for B cells and T cells as indicated based on CD3 and CD20 staining. b, Fold change (log2-transformation) in expression of various markers of T cell activation and response in TLS-associated T cells as compared to T cells found outside the TLS per individual slide. Data show individual TLS ROI values divided by the average non-TLS value of that slide. Increased expression in the context of TLSs is represented by shaded pink box (>0). c, Average log2-transformed fold change of expression for TLS-associated T cells as compared to non-associated T cells. Individual dots represent individual patients/slides as indicated. Data show the average log2-transformed count per TLS ROI value minus the average log2-transformed count per non-TLS ROI value per slide for each protein queried. For b and c, increased expression in the context of TLSs is represented by shaded pink box (>0). Median and interquartile range are indicated. Error bars indicate 95% confidence intervals. d, Levels of Ki67 protein expression in B cell masks of non-TLSs and TLS ROIs by individual patient as indicated. Counts are represented as signal-to-noise ratios of Ki67 compared to geometric means of isotype controls. Median and interquartile range are indicated. Error bars indicated 95% confidence ratios, and P values were determined by Student’s t-test. For a–d, the number of ROIs analysed for each patient are 11 for patient 1, 12 for patient 2, 12 for patient 10, 7 for patient 17 and 7 for patient 19.
Extended Data Fig. 8 BCR analyses of intratumoral B cells in patients with advanced melanoma before and after treatment with neoadjuvant ICB.
a, Clonal counts for all identified clonotypes for both IgH and IgL after treatment with ICB. Patients are grouped as responders and non-responders and identified as indicated in which each bar represents individual patient. b, Clonal proportion for all identified clonotypes for both IgH and IgL for baseline samples further evaluated in Fig. 3a and on-treatment samples in a. Patients are grouped as responders and non-responders and identified as indicated in which each bar represents individual patient. c, d, Summed expression of top five clonotypes in normalized read counts (c) and BCR repertoire diversity (d) for responders and non-responders for both IgH and IgL at baseline (n = 11 NR and 10 R for IgH and IgL) and on-treatment (n = 10 NR and 9 R for IgH and n = 11 NR and 9 R for IgL). Box plot shows median and interquartile range. P values determined by two-sided Mann–Whitney U-test.
Extended Data Fig. 9 Single-cell RNA-seq analysis reveals unique clusters of B cells associated with response to ICB.
a, Scatter plots comparing the percentage of CD45+ cells staining positive for CD19+ (B cells) as indicated between responder (n = 17) and non-responder (n = 31) samples with all time points combined or stratified by pre- and post-treatment as indicated. Data are median and interquartile range. P values were determined by two-sided Mann–Whitney U-test. b, Heat map displaying scaled expression values (log2(TPM + 1) of discriminative genes from all B cells between responder (blue) and non-responder (red) samples. Top marker genes are shown for each group. c, Heat map showing scaled expression values (log2(TPM + 1)) of discriminative gene sets per cluster as defined in Fig. 3c. A list of representative genes is shown per cluster next to the left margin. For both heat maps, colour scheme is based on z-scores from −2.5 (blue) to 2.5 (yellow).
Extended Data Fig. 10 Mass cytometry reveals significant differences in Bcell populations between responder and non-responder tumours.
a, Pie charts representing composition of individual tumour and peripheral blood samples for patients with melanoma treated with ICB used in all analyses for mass cytometry. Matched patient samples are located directly beneath one another. Samples from patients with lymph node or non-lymph-node metastases as indicated. Cell types as indicated. Asterisk indicates samples included in t-SNE plots and pie charts in c, Fig. 3d–f, and phenographs in Extended Data Fig. 11. b, Scatter plots demonstrating quantification of different peripheral blood and intratumoral B cell phenotypes. Median and interquartile range are shown. All samples are represented in b (for tumour, n = 7 R and 3 NR and, for peripheral blood, n = 4 R and 4 NR). P values were determined by one-sided Mann–Whitney U-test. c, t-SNE plots demonstrating intratumoral B cell phenotypes from the neoadjuvant ICB trial in patients with advanced melanoma grouped by response and including further breakdown of memory cell subtypes and germinal centre B cells. Plots represent combined analyses of tumours ran simultaneously with the peripheral blood samples (n = 5 R and 3 NR) and include baseline and on-treatment samples as detailed in Supplementary 31. d, Quantification of B cell subtypes in tumour from mass cytometric analyses in responders and non-responder from all tumours (n = 7 R and 3 NR). Median and interquartile range are shown. P values were determined by one-sided Mann–Whitney U-test.
Extended Data Fig. 11 Surface expression of markers analysed by mass cytometry.
a, Individual phenographs for surface expression of each marker analysed as indicated. These data represent combined tumour and peripheral blood samples from patients with melanoma treated with ICB ran together (8 tumour n = 5 R and n = 3 NR and 8 peripheral blood samples n = 4 R and n = 4 NR), thus eliminating batch effect. b, Percentage of CD45+CD19+ cells by tissue type—peripheral blood versus tumour—that are positive for each of the surface markers indicated. c, Percentage of CD45+CD19+ cells in tumour by response—responder versus non-responder—that are positive for each of the surface markers indicated. For b and c, all samples are represented (for tumour, n = 7 R and 3 NR and, for peripheral blood, n = 4 R and n = 4 NR). Error bars indicate median and interquartile range. P values were determined by two-sided Mann–Whitney U-test.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1, 5, 7-8, 10, 14, 18, 22-23, 26, 28-32.
Supplementary Tables
This file contains Supplementary Tables 2-4, 6, 9, 11-13, 15-17, 19-21, 24-25, and 27.
Supplementary Information
This file contains a full Supplementary Tables Guide for tables 1-32.
Rights and permissions
About this article
Cite this article
Helmink, B.A., Reddy, S.M., Gao, J. et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 577, 549–555 (2020). https://doi.org/10.1038/s41586-019-1922-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1922-8