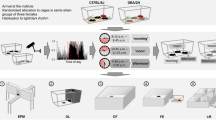
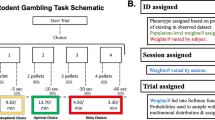
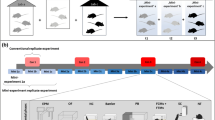
Abstract
Low statistical power reduces the reliability of animal research; yet, increasing sample sizes to increase statistical power is problematic for both ethical and practical reasons. We present an alternative solution using Bayesian priors based on historical control data, which capitalizes on the observation that control groups in general are expected to be similar to each other. In a simulation study, we show that including data from control groups of previous studies could halve the minimum sample size required to reach the canonical 80% power or increase power when using the same number of animals. We validated the approach on a dataset based on seven independent rodent studies on the cognitive effects of early-life adversity. We present an open-source tool, RePAIR, that can be widely used to apply this approach and increase statistical power, thereby improving the reliability of animal experiments.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The data that support the findings of the current study can be downloaded from https://osf.io/wvs7m/.
Code availability
All code used in this manuscript is available at https://osf.io/wvs7m/.
References
Ioannidis, J. P. A. Why most published research findings are false. PLoS Med. 2, e124 (2005).
Button, K. S. et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376 (2013).
Smaldino, P. E. & McElreath, R. The natural selection of bad science. R. Soc. Open Sci. 3, 160384 (2016).
Munafò, M. R. et al. A manifesto for reproducible science. Nat. Hum. Behav. 1, 0021 (2017).
Crabbe, J. C., Wahlsten, D. & Dudek, B. C. Genetics of mouse behavior: interactions with laboratory environment. Science 284, 1670–1672 (1999).
Prinz, F., Schlange, T. & Asadullah, K. Believe it or not: how much can we rely on published data on potential drug targets? Nat. Rev. Drug Discov. 10, 712–713 (2011).
Voelkl, B. et al. Reproducibility of animal research in light of biological variation. Nat. Rev. Neurosci. 21, 384–393 (2020).
Macleod, M. & Mohan, S. Reproducibility and rigor in animal-based research. ILAR J. 60, 17–23 (2019).
Bonapersona, V. et al. The behavioral phenotype of early life adversity: a 3-level meta-analysis of rodent studies. Neurosci. Biobehav. Rev. 102, 299–307 (2019).
Gurevitch, J., Koricheva, J., Nakagawa, S. & Stewart, G. Meta-analysis and the science of research synthesis. Nature 555, 175–182 (2018).
Richter, S. H., Garner, J. P. & Würbel, H. Environmental standardization: cure or cause of poor reproducibility in animal experiments? Nat. Methods 6, 257–261 (2009).
Karp, N. A. Reproducible preclinical research—is embracing variability the answer? PLoS Biol. 16, e2005413 (2018).
Richter, S. H. et al. Effect of population heterogenization on the reproducibility of mouse behavior: a multi-laboratory study. PLoS ONE 6, e16461 (2011).
Kramer, M. & Font, E. Reducing sample size in experiments with animals: historical controls and related strategies. Biol. Rev. Philos. Soc. 92, 431–445 (2017).
Brakenhoff, T., Roes, K. & Nikolakopoulos, S. Bayesian sample size re-estimation using power priors. Stat. Methods Med. Res. 28, 1664–1675 (2018).
Spiegelhalter, D. J., Abrams, K. R. & Myles, J. P. Bayesian Approaches to Clinical Trials and Health-Care Evaluation 1 (Wiley, 2004).
Galwey, N. W. Supplementation of a clinical trial by historical control data: is the prospect of dynamic borrowing an illusion? Stat. Med. 36, 899–916 (2017).
Mutsvari, T., Tytgat, D. & Walley, R. Addressing potential prior-data conflict when using informative priors in proof-of-concept studies. Pharm. Stat. 15, 28–36 (2016).
Walley, R. et al. Using Bayesian analysis in repeated preclinical in vivo studies for a more effective use of animals. Pharm. Stat. 15, 277–285 (2016).
Nord, C. L., Valton, V., Wood, J. & Roiser, J. P. Power-up: a reanalysis of ‘power failure’ in neuroscience using mixture modeling. J. Neurosci. 37, 8051–8061 (2017).
Cohen, J. Statistical Power Analysis for the Behavioral Sciences (Lawrence Erlbaum Associates, 1977).
Ibrahim, J. G. & Chen, M. H. Power prior distributions for regression models. Stat. Sci. 15, 46–60 (2000).
Rice, C. J., Sandman, C. A., Lenjavi, M. R. & Baram, T. Z. A novel mouse model for acute and long-lasting consequences of early life stress. Endocrinology 149, 4892–4900 (2008).
Percie du Sert, N. et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 18, e3000410 (2020).
Ioannidis, J. P. A. Why most discovered true associations are inflated. Epidemiology 19, 640–648 (2008).
Rubin, E. J. & Fortune, S. M. Misunderstanding the goals of animal research. BMJ 360, 29321149 (2018).
Gelman, A. Objections to Bayesian statistics. Bayesian Anal. 3, 445–450 (2008).
Viele, K. et al. Use of historical control data for assessing treatment effects in clinical trials. Pharm. Stat. 13, 41–54 (2014).
Neuenschwander, B., Capkun-Niggli, G., Branson, M. & Spiegelhalter, D. J. Summarizing historical information on controls in clinical trials. Clin. Trials 7, 5–18 (2010).
O’Hagan, A. Expert knowledge elicitation: subjective but scientific. Am. Stat. 73, 69–81 (2019).
van der Naald, M., Wenker, S., Doevendans, P. A., Wever, K. E. & Chamuleau, S. A. J. Publication rate in preclinical research: a plea for preregistration. BMJ Open Sci. 4, e100051 (2020).
Du Sert, N. P. et al. The experimental design assistant. Nat. Methods 14, 1024–1025 (2017).
Crabbe, J. C. & Phillips, T. J. Mother nature meets mother nurture. Nat. Neurosci. 6, 440–442 (2003).
Kafkafi, N. et al. Addressing reproducibility in single-laboratory phenotyping experiments. Nat. Methods 14, 462–464 (2017).
Shansky, R. M. Are hormones a ‘female problem’ for animal research? Science 364, 825–826 (2019).
Bonapersona, V., Joëls, M. & Sarabdjitsingh, R. A. Effects of early life stress on biochemical indicators of the dopaminergic system: a 3 level meta-analysis of rodent studies. Neurosci. Biobehav. Rev. 95, 1–16 (2018).
Wickham, H. et al. Welcome to the Tidyverse. J. Open Source Softw. 4, 1686 (2019).
Chang, W., Cheng, J., Allaire, J., Xie, Y. & McPherson, J. shiny: Web Application Framework for R. R Package version 1.5.0 https://CRAN.R-project.org/package=shiny (2020).
Ekstrøm, C. T. MESS: Miscellaneous Esoteric Statistical Scripts. R package version 0.5.6 https://CRAN.R-project.org/package=MESS (2019).
Faul, F., Erdfelder, E., Lang, A. G. & Buchner, A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 39, 175–191 (2007).
Lenth, R. V. Some practical guidelines for effective sample size determination. Am. Stat. 55, 187–193 (2001).
Gelman, A., Carlin, J. B., Stern, H. S. & Rubin, D. B. Bayesian Data Analysis. (Chapman & Hall, 1995).
Acknowledgements
We thank J. Knop and M. Sep for helpful discussions and R. de Kloet for critically reviewing the manuscript. This work was supported by the Consortium of Individual Development (CID), which is funded by the Gravitation program of the Dutch Ministry of Education, Culture and Science and the Netherlands Organization for Scientific Research (NWO grant no. 024.001.003).
Author information
Authors and Affiliations
Consortia
Contributions
V.B. contributed to conceptualization, data curation, analysis, investigation, methodology, software, visualization and writing the manuscript; H.H. contributed to conceptualization, analysis, methodology, supervision and reviewing and editing the manuscript; members of the RELACS consortium provided the data; R.A.S. contributed to conceptualization, project administration, supervision and editing and reviewing the manuscript; M.J. contributed to conceptualization, funding acquisition, project administration, supervision and writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Stanley Lazic, Malcolm MacLeod, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–4, Supplementary Fig. 1 and Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Bonapersona, V., Hoijtink, H., RELACS Consortium. et al. Increasing the statistical power of animal experiments with historical control data. Nat Neurosci 24, 470–477 (2021). https://doi.org/10.1038/s41593-020-00792-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00792-3
This article is cited by
-
Challenging current scientific practice: how a shift in research methodology could reduce animal use
Lab Animal (2024)
-
A minimal metadata set (MNMS) to repurpose nonclinical in vivo data for biomedical research
Lab Animal (2024)
-
Statistical power and false positive rates for interdependent outcomes are strongly influenced by test type: Implications for behavioral neuroscience
Neuropsychopharmacology (2023)
-
“The wrong tools for the right job”: a critical meta-analysis of traditional tests to assess behavioural impacts of maternal separation
Psychopharmacology (2023)
-
Psychiatric-Like Impairments in Mouse Models of Spinocerebellar Ataxias
The Cerebellum (2022)