Abstract
Animals coexist in commensal, pathogenic or mutualistic relationships with complex communities of diverse organisms, including microorganisms1. Some bacteria produce bioactive neurotransmitters that have previously been proposed to modulate nervous system activity and behaviours of their hosts2,3. However, the mechanistic basis of this microbiota–brain signalling and its physiological relevance are largely unknown. Here we show that in Caenorhabditis elegans, the neuromodulator tyramine produced by commensal Providencia bacteria, which colonize the gut, bypasses the requirement for host tyramine biosynthesis and manipulates a host sensory decision. Bacterially produced tyramine is probably converted to octopamine by the host tyramine β-hydroxylase enzyme. Octopamine, in turn, targets the OCTR-1 octopamine receptor on ASH nociceptive neurons to modulate an aversive olfactory response. We identify the genes that are required for tyramine biosynthesis in Providencia, and show that these genes are necessary for the modulation of host behaviour. We further find that C. elegans colonized by Providencia preferentially select these bacteria in food choice assays, and that this selection bias requires bacterially produced tyramine and host octopamine signalling. Our results demonstrate that a neurotransmitter produced by gut bacteria mimics the functions of the cognate host molecule to override host control of a sensory decision, and thereby promotes fitness of both the host and the microorganism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data necessary to reproduce these analyses are available as Source Data and/or at https://github.com/SenguptaLab/ProvidenciaChemo.git. Raw data from HPLC–MS experiments are available upon request (owing to the large file sizes).
Code availability
All statistical analysis code necessary to reproduce these analyses are available at https://github.com/SenguptaLab/ProvidenciaChemo.git.
References
Douglas, A. E. Fundamentals of Microbiome Science: How Microbes Shape Animal Biology (Princeton Univ. Press, 2018).
Guo, R., Chen, L.-H., Xing, C. & Liu, T. Pain regulation by gut microbiota: molecular mechanisms and therapeutic potential. Br. J. Anaesth. 123, 637–654 (2019).
Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 1693 (Pt B), 128–133 (2018).
Zhang, J., Holdorf, A. D. & Walhout, A. J. C. elegans and its bacterial diet as a model for systems-level understanding of host–microbiota interactions. Curr. Opin. Biotechnol. 46, 74–80 (2017).
Schulenburg, H. & Félix, M.-A. The natural biotic environment of Caenorhabditis elegans. Genetics 206, 55–86 (2017).
Meisel, J. D. & Kim, D. H. Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 35, 465–470 (2014).
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M.-A. & Ruvkun, G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl Acad. Sci. USA 113, E3941–E3949 (2016).
Bargmann, C. I., Hartwieg, E. & Horvitz, H. R. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527 (1993).
Song, B.-M., Faumont, S., Lockery, S. & Avery, L. Recognition of familiar food activates feeding via an endocrine serotonin signal in Caenorhabditis elegans. eLife 2, e00329 (2013).
Chao, M. Y., Komatsu, H., Fukuto, H. S., Dionne, H. M. & Hart, A. C. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc. Natl Acad. Sci. USA 101, 15512–15517 (2004).
Liang, B., Moussaif, M., Kuan, C.-J., Gargus, J. J. & Sze, J. Y. Serotonin targets the DAF-16/FOXO signaling pathway to modulate stress responses. Cell Metab. 4, 429–440 (2006).
Entchev, E. V. et al. A gene-expression-based neural code for food abundance that modulates lifespan. eLife 4, e06259 (2015).
Avery, L. & Shtonda, B. B. Food transport in the C. elegans pharynx. J. Exp. Biol. 206, 2441–2457 (2003).
Avery, L. The genetics of feeding in Caenorhabditis elegans. Genetics 133, 897–917 (1993).
Berg, M. et al. Assembly of the Caenorhabditis elegans gut microbiota from diverse soil microbial environments. ISME J. 10, 1998–2009 (2016).
Dirksen, P. et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host–microbiome model. BMC Biol. 14, 38 (2016).
Tan, M.-W., Mahajan-Miklos, S. & Ausubel, F. M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl Acad. Sci. USA 96, 715–720 (1999).
Irazoqui, J. E. et al. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6, e1000982 (2010).
Wragg, R. T. et al. Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. J. Neurosci. 27, 13402–13412 (2007).
Mills, H. et al. Monoamines and neuropeptides interact to inhibit aversive behaviour in Caenorhabditis elegans. EMBO J. 31, 667–678 (2012).
Harris, G. et al. The monoaminergic modulation of sensory-mediated aversive responses in Caenorhabditis elegans requires glutamatergic/peptidergic cotransmission. J. Neurosci. 30, 7889–7899 (2010).
Ezak, M. J. & Ferkey, D. M. The C. elegans D2-like dopamine receptor DOP-3 decreases behavioral sensitivity to the olfactory stimulus 1-octanol. PLoS ONE 5, e9487 (2010).
Ezcurra, M., Tanizawa, Y., Swoboda, P. & Schafer, W. R. Food sensitizes C. elegans avoidance behaviours through acute dopamine signalling. EMBO J. 30, 1110–1122 (2011).
Alkema, M. J., Hunter-Ensor, M., Ringstad, N. & Horvitz, H. R. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260 (2005).
Lints, R. & Emmons, S. W. Patterning of dopaminergic neurotransmitter identity among Caenorhabditis elegans ray sensory neurons by a TGFβ family signaling pathway and a Hox gene. Development 126, 5819–5831 (1999).
Sze, J. Y., Victor, M., Loer, C., Shi, Y. & Ruvkun, G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564 (2000).
Troemel, E. R., Chou, J. H., Dwyer, N. D., Colbert, H. A. & Bargmann, C. I. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell 83, 207–218 (1995).
Artyukhin, A. B. et al. Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J. Biol. Chem. 288, 18778–18783 (2013).
Pugin, B. et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 28, 1353881 (2017).
Barbieri, F., Montanari, C., Gardini, F. & Tabanelli, G. Biogenic amine production by lactic acid bacteria: a review. Foods 8, 17 (2019).
Marcobal, A., Martín-Alvarez, P. J., Moreno-Arribas, M. V. & Muñoz, R. A multifactorial design for studying factors influencing growth and tyramine production of the lactic acid bacteria Lactobacillus brevis CECT 4669 and Enterococcus faecium BIFI-58. Res. Microbiol. 157, 417–424 (2006).
Duerr, J. S. et al. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J. Neurosci. 19, 72–84 (1999).
Sandmeier, E., Hale, T. I. & Christen, P. Multiple evolutionary origin of pyridoxal-5′-phosphate-dependent amino acid decarboxylases. Eur. J. Biochem. 221, 997–1002 (1994).
Connil, N. et al. Identification of the Enterococcus faecalis tyrosine decarboxylase operon involved in tyramine production. Appl. Environ. Microbiol. 68, 3537–3544 (2002).
Linares, D. M., Fernández, M., Martín, M. C. & Alvarez, M. A. Tyramine biosynthesis in Enterococcus durans is transcriptionally regulated by the extracellular pH and tyrosine concentration. Microb. Biotechnol. 2, 625–633 (2009).
Zhu, H. et al. Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding. Sci. Rep. 6, 27779 (2016).
Quick, M. et al. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter:sodium symporter from Fusobacterium nucleatum. J. Biol. Chem. 281, 26444–26454 (2006).
Collins, K. M. et al. Activity of the C. elegans egg-laying behavior circuit is controlled by competing activation and feedback inhibition. eLife 5, e21126 (2016).
Rex, E. et al. TYRA-2 (F01E11.5): a Caenorhabditis elegans tyramine receptor expressed in the MC and NSM pharyngeal neurons. J. Neurochem. 94, 181–191 (2005).
Sun, J., Singh, V., Kajino-Sakamoto, R. & Aballay, A. Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332, 729–732 (2011).
Elgaali, H. et al. Comparison of long-chain alcohols and other volatile compounds emitted from food-borne and related Gram positive and Gram negative bacteria. J. Basic Microbiol. 42, 373–380 (2002).
Worthy, S. E. et al. Identification of attractive odorants released by preferred bacterial food found in the natural habitats of C. elegans. PLoS ONE 13, e0201158 (2018).
Yoshida, K. et al. Odour concentration-dependent olfactory preference change in C. elegans. Nat. Commun. 3, 739 (2012).
Zhang, F. et al. Caenorhabditis elegans as a model for microbiome research. Front. Microbiol. 8, 485 (2017).
Leitão-Gonçalves, R. et al. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 15, e2000862 (2017).
Pais, I. S., Valente, R. S., Sporniak, M. & Teixeira, L. Drosophila melanogaster establishes a species-specific mutualistic interaction with stable gut-colonizing bacteria. PLoS Biol. 16, e2005710 (2018).
Henriques, S. F. et al. Metabolic cross-feeding allows a gut microbial community to overcome detrimental diets and alter host behaviour. Preprint at https://www.biorxiv.org/content/10.1101/821892v1 (2019).
Breton, J. et al. Gut commensal E. coli proteins activate host satiety pathways following nutrient-induced bacterial growth. Cell Metab. 23, 324–334 (2016).
Fetissov, S. O. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nat. Rev. Endocrinol. 13, 11–25 (2017).
Blomfield, I. C., Vaughn, V., Rest, R. F. & Eisenstein, B. I. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5, 1447–1457 (1991).
Jiang, Y. et al. Multigene editing in the Escherichia coli genome via the CRISPR–Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015).
Marx, C. J. Development of a broad-host-range sacB-based vector for unmarked allelic exchange. BMC Res. Notes 1, 1 (2008).
Barbier, M. & Damron, F. H. Rainbow vectors for broad-range bacterial fluorescence labeling. PLoS ONE 11, e0146827 (2016).
Alegado, R. A. & Tan, M.-W. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell. Microbiol. 10, 1259–1273 (2008).
Troemel, E. R., Kimmel, B. E. & Bargmann, C. I. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169 (1997).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017).
Hunt, M. et al. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16, 294 (2015).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Lassmann, T. & Sonnhammer, E. L. L. Kalign—an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics 6, 298 (2005).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27, 1164–1165 (2011).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
R Core Team. R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2019).
RStudio Team. RStudio: integrated development for R (RStudio, 2019).
Bates, D., Maechler, M., Bolker, B. & Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015).
Lenth, R. emmeans: estimated marginal means, aka least-squares means. R package v.1.4.5, https://CRAN.R-project.org/package=emmeans (2020).
Halekoh, U. & Højsgaard, S. A Kenward–Roger approximation and parametric bootstrap methods for tests in linear mixed models – the R package pbkrtest. J. Stat. Softw. 59, 1–30 (2014).
Kuznetsova, A., Brockhoff, P. B. & Christensen, R. H. B. lmerTest package: tests in linear mixed effects models. J. Stat. Softw. 82, 1–26 (2017).
Fox, J. & Weisberg, S. An R Companion to Applied Regression 3rd edn (Sage, 2019).
Goodrich, B., Gabry, J., Ali, I. & Brilleman, S. rstanarm: Bayesian applied regression modeling via Stan, https://mc-stan.org/rstanarm (2020).
Plummer, M., Best, N., Cowles, K. & Vines, K. CODA: convergence diagnosis and output analysis for MCMC. R News 6, 7–11 (2006).
Gabry, J., Simpson, D., Vehtari, A., Betancourt, M. & Gelman, A. Visualization in Bayesian workflow. J. R. Stat. Soc. A. 182, 389–402 (2019).
Kay, M. tidybayes: tidy data and geoms for Bayesian models, https://doi.org/10.5281/zenodo.1308151 (2020).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S 4th edn (Springer, 2002).
Helf, M. Metaboseek: an interactive, browser-based tool to analyze your mass spectrometry data, https://doi.org/10.5281/zenodo.3360087 (2019).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Acknowledgements
We thank R. Komuniecki for the octr-1 cDNA; M. Alkema for the tbh-1 promoter; multiple members of the C. elegans community for bacterial strains (listed in Supplementary Table 2); the Caenorhabditis Genetics Center for C. elegans strains; the Broad Institute for bacterial genome sequencing; M. Helf for assistance with the Metaboseek software; J. Yu and H. Le for synthesis of N-succinyl neurotransmitter standards; S. Lovett, L. Laranjo, M. Alkema and the laboratory of P.S. for advice; and C. Bargmann, O. Hobert and the laboratory of P.S. for comments on the manuscript. This work was partly supported by the NIH (T32 NS007292 and F32 DC013711 to M.P.O.; R01 GM088290 and R35 GM131877 to F.C.S.; and R35 GM122463 and R21 NS101702 to P.S.) and the NSF (IOS 1655118 to P.S.).
Author information
Authors and Affiliations
Contributions
M.P.O., B.W.F., F.C.S. and P.S. designed experiments, interpreted results and wrote the paper with input from all authors. M.P.O. and P.-H.C. conducted long-range chemotaxis behavioural experiments and analysed results. B.W.F. conducted HPLC–MS experiments and analysed results. M.P.O. conducted and analysed results from all additional experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks William Schafer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Octanol modulation by Providencia requires ingestion of bacteria and is not mediated by nutritive cues.
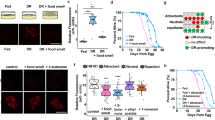
a, Cartoon and data from long-range chemotaxis assays of C. elegans grown on the indicated bacterial strains to the indicated attractive odours. Dots, chemotaxis index from single assays of approximately 100 worms. Horizontal line, median; errors, 1st and 3rd quartiles. Numbers in parentheses, number of assays performed over at least three days. b, Cartoon and data from osmotic ring avoidance assay. Dots, single assays of 10 worms. Numbers in parentheses, number of assays over at least three independent days. Y axis is the proportion of worms leaving an osmotic ring barrier of 8 M glycerol after 10 min. P value represents difference of means relative to worms grown on JUb39 from a GLMM. Errors are s.e.m. Grey thin and thick vertical bars, Bayesian 95% and 66% credible intervals, respectively. c, Isolation of nematode-associated bacteria. Nematodes were isolated from residential compost in Massachusetts (USA). Worms were allowed to crawl onto NGM plates from which they were picked to clean plates. Resulting bacterial colonies were isolated, grown on LB medium and characterized via 16S rRNA sequencing. d, Expression of a tph-1p::gfp fluorescent reporter in indicated head neurons of young adult worms grown on either OP50 or JUb39. Dots, mean fluorescence of the soma of single neurons. Horizontal bar is mean; errors are s.e.m. Grey thin and thick vertical bars, Bayesian 95% and 66% credible intervals, respectively. P values are from two-way ANOVA. e, f, Modulation index of worms grown on the indicated bacterial strains to 100% octanol, under the shown conditions. Worms were exposed to the indicated bacteria on the plate lid (e) for one generation, or to NGM control or bacteria-conditioned NGM (f) for 2 h before the assay. Numbers in parentheses, independent experiments over 2 d with approximately 100 worms each. Values are shown on a log-odds (logit) scale and are normalized to the values of wild-type worms grown on OP50 for each day, indicated with a grey dashed line. Positive numbers indicate reduced avoidance of octanol. Errors are s.e.m. Grey thin and thick vertical bars, Bayesian 95% and 66% credible intervals, respectively. P values between the indicated conditions are post hoc comparisons from a GLMM, with Tukey-type multivariate t adjustment for f. g, Survival analysis of worms grown on the indicated strains. Dots, average proportion of surviving worms from 12 plates of 20 worms each on the indicated days. Error bars are s.e.m. Shaded blue and grey curves indicate 95% confidence intervals derived from 1,000 bootstrap Gompertz function nonlinear least squares (nls) fits of the indicated data. Top blue and grey distributions show bootstrapped median survival of the indicated strains.
Extended Data Fig. 2 Detection of N-succinyl tyramine.
a, b, Major fragmentation reactions and resulting fragment ions are indicated in mass spectra of N-succinyl tyramine obtained in ESI− (a) and ESI+ (b) HPLC–MS2. Representative data are shown from at least three biologically independent experiments.
Extended Data Fig. 3 Complementation of tyramine-containing metabolites in tdc-1-mutant worms via Providencia AADCs.
a, Overview of MS2 network obtained in positive ion mode. Each node represents a unique feature, and edges between nodes indicate similarity between MS2 spectra. Red circle highlights the subnetwork containing the majority (16 out of 24 features, representing 10 out of 15 differential compounds) of differential tyramine-containing metabolites. This subnetwork contains glycosylated tyramine derivatives; N-succinyl tyramine is represented by a node in a different sub-network (red arrow). b, Magnified view of subnetwork highlighted in a. The tyramine-containing features restored upon growth of tdc-1-mutant worms on wild-type JUb39, and abolished when grown on ΔtyrDC::cmR ΔadcA JUb39, are highlighted in light red. MS2 spectra for two example compounds at m/z 499.1474 and m/z 604.1689 (circled in magenta) are shown in Extended Data Figs. 4 and 5, respectively.
Extended Data Fig. 4 MS2 analysis and quantification of the tyramine-containing metabolite m/z 499.1474.
a, Major fragmentation reactions of m/z 499.1474 and resulting fragment ions. The putative structure shown is based on fragmentation and stable-isotope incorporation. The stereochemistry and exact substitution pattern are unknown. b, c, MS2 spectra obtained in ESI− (b) and ESI+ (c) mode. Representative data are shown from at least three biologically independent experiments. d, Quantification of m/z 499.1474 in wild-type and tdc-1-mutant worms fed the indicated bacterial strains as determined by positive-ion ESI+ HPLC–MS. Dots, independent samples from n = 3 experiments. Errors are s.e.m.
Extended Data Fig. 5 MS2 analysis and quantification of the tyramine-containing metabolite m/z 604.1689.
a, Major fragmentation reactions of m/z 604.1689 and resulting fragment ions. The stereochemistry and exact substitution pattern are unknown. The structure shown is based on fragmentation and stable-isotope incorporation. b, c, MS2 spectra obtained in negative-ion (b) and positive-ion (c) mode. Representative data are shown from at least three biologically independent experiments. d, Quantification of m/z 604.1689 in wild-type and tdc-1-mutant worms fed the indicated bacterial strains as determined by positive-ion ESI HPLC–MS. Dots, independent samples from n = 3 experiments. Errors are s.e.m.
Extended Data Fig. 6 Quantification of N-acetyl serotonin in worms fed the indicated bacteria, or in bacterial cultures alone, as determined by ESI+ HPLC–MS.
Dots, independent samples from n = 3 experiments. Errors are s.e.m.
Extended Data Fig. 7 l-Tyr supplementation enhances octanol modulation.
a, b, Reversal response times of worms of the indicated genotypes grown on the indicated bacteria in control conditions (a) or supplemented with 0.5% l-Tyr (a, b) to 100% octanol using SOS assays. Dots, response time of single worms. Y axis is log10-scaled for these log-normally distributed data, and normalized to the indicated control group for each experimental day. Numbers in parentheses, number of worms tested in assays over at least three independent days. Box plot, median and quartiles; whiskers, data range (excluding outliers). Grey thin and thick vertical bars, Bayesian 95% and 66% credible intervals for the difference of means, respectively. P values indicating comparisons of means relative to the OP50 control for each of the conditions are from an LMM with Tukey-type multivariate t adjustment. P value in red indicates Wald F-statistic (a) or Wald t-statistic (b) for the effect of l-Tyr supplementation (a) or genotype (b) on the magnitude of the JUb39 effect. Wild-type data in b are also shown in Fig. 2g.
Extended Data Fig. 8 Phylogenetic analysis of group-II decarboxylase genes in Gammaproteobacteria.
a, Neighbour-joining unrooted tree based on sequences identified via a BLAST search using E. faecalis TyrDC and C. elegans TDC-1. The initial tree indicates three major groups. Representative enzymes and operon structures for each group are indicated by coloured boxes. b, Bootstrapped maximum likelihood phylogeny using PhyML and Phylomizer pipeline. Maximum of two highly similar sequences per genus were included after each BLAST search. Genera are indicated to the right. Numbers on branch-points matching this tree out of 100 bootstrap replicates are indicated at values >60. Group representatives from a are indicated in corresponding colours. Providencia and C. elegans sequences discussed in this Article are indicated in bold. Accession numbers and BLAST metrics are listed in Supplementary Table 1. c, Presence of tyrDC and adcA among complete genomes in Gammaproteobacteria. Linked boxes indicate organization in an operon. Hatched shading indicates variable presence among genera. Coloured triangles indicate taxa of interest. d, Homology-based model of the TyrDC catalytic domain in Providencia based on the Lactobacillus TyrDC crystal structure36 using SWISS-MODEL (https://swissmodel.expasy.org). Residues in magenta, green and yellow are from Lactobacillus TyrDC, JUb39 TyrDC and JUb39 AdcA, respectively. Pyridoxyl phosphate (PLP) is depicted in red and l-Tyr (manually docked for illustration) is indicated in light blue. Position of A600 and S58636 in JUb39 TyrDC and Lactobacillus TyrDC, respectively, are indicated.
Extended Data Fig. 9 Disruption of JUb39 tyramine production or host octopamine receptor signalling affects octanol modulation without altering intestinal bacterial cell numbers.
a, Presence of mCherry-expressing bacteria in the posterior intestines of young adult wild-type or octr-1-mutant worms. Bars show proportion of worms with the indicated distribution of bacterial cells present in worms grown on the bacteria indicated. Numbers in parentheses, number of worms. P value is derived from an ordinal regression. b, Reversal response latency of worms of the indicated genotypes grown on the bacteria indicated in control conditions of NGM + 0.5% l-Tyr to 100% octanol using SOS assays. Dots, response time of single worms. Y axis is log10-scaled for these log-normally distributed data, and normalized to the indicated control group for each experimental day. Numbers in parentheses, number of worms tested in assays over at least three independent days. Box plot, median and quartiles; whiskers, data range (excluding outliers). Grey thin and thick vertical bars, Bayesian 95% and 66% credible intervals for the difference of means, respectively. P values between indicated conditions are from an LMM with Tukey-type multivariate t adjustment. P values in red indicate Wald t-statistic representing the genotype × food interaction effect relative to wild type.
Extended Data Fig. 10 Providencia-derived tyramine complements the egg-laying defects of tdc-1-mutant C. elegans.
Quantification of the age of eggs in utero of worms of the indicated genotypes, grown on the indicated bacterial strains. Dots, proportion of eggs at or older than the four-cell stage in individual worms. Numbers in parentheses, total number of worms scored in three independent experiments. Box plot, median and quartiles; whiskers, data range (excluding outliers). P values between the indicated conditions are from a binomial generalized linear mixed effects regression using a logit link function, with a post hoc Tukey correction for multiple comparisons.
Supplementary information
Supplementary Table 1
| Group II decarboxylase-encoding genes identified in Gammaproteobacteria. Genes were identified using a hierarchical tblastn approach (see Methods). Blast query sequence, query group, and the presence of putative transporter-encoding genes are shown in the indicated columns.
Supplementary Information Table 2
| Strains used in this work.
Source data
Rights and permissions
About this article
Cite this article
O’Donnell, M.P., Fox, B.W., Chao, PH. et al. A neurotransmitter produced by gut bacteria modulates host sensory behaviour. Nature 583, 415–420 (2020). https://doi.org/10.1038/s41586-020-2395-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2395-5
This article is cited by
-
Carbon-nanotube field-effect transistors for resolving single-molecule aptamer–ligand binding kinetics
Nature Nanotechnology (2024)
-
Larval microbiota primes the Drosophila adult gustatory response
Nature Communications (2024)
-
Vitamin B12 produced by gut bacteria modulates cholinergic signalling
Nature Cell Biology (2024)
-
Repurposing degradation pathways for modular metabolite biosynthesis in nematodes
Nature Chemical Biology (2023)
-
Parallel pathways for serotonin biosynthesis and metabolism in C. elegans
Nature Chemical Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.