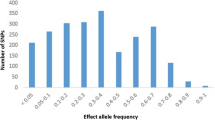
Abstract
Type 2 diabetes has been reproducibly clustered into five subtypes with different disease progression and risk of complications; however, etiological differences are unknown. We used genome-wide association and genetic risk score (GRS) analysis to compare the underlying genetic drivers. Individuals from the Swedish ANDIS (All New Diabetics In Scania) study were compared to individuals without diabetes; the Finnish DIREVA (Diabetes register in Vasa) and Botnia studies were used for replication. We show that subtypes differ with regard to family history of diabetes and association with GRS for diabetes-related traits. The severe insulin-resistant subtype was uniquely associated with GRS for fasting insulin but not with variants in the TCF7L2 locus or GRS reflecting insulin secretion. Further, an SNP (rs10824307) near LRMDA was uniquely associated with mild obesity-related diabetes. Therefore, we conclude that the subtypes have partially distinct genetic backgrounds indicating etiological differences.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others

Data availability
Individual-level data are not publicly available due to ethical and legal restrictions related to the Swedish Biobanks in Medical Care Act (2002:297) and Personal Data Act (1998:204), the European Union’s General Data Protection Regulation 2016/679 and the Data Protection Act 2018:218. Supporting data are available from the corresponding author upon reasonable request but access to data must be granted by the ANDIS and MDC steering committees. Summary statistics are available in the GWAS Catalog (www.ebi.ac.uk/gwas/) under accession no. GCST90026412-7. SNPs and the weights for GRS are available in the supplementary tables and in the Polygenic Score Catalog (www.pgscatalog.org) under IDs PGS000832-64 and PGS000024.
Code availability
No custom code was used for the analysis of data.
References
Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 6, 361–369 (2018).
Zaharia, O. P. et al. Risk of diabetes-associated diseases in subgroups of patients with recent-onset diabetes: a 5-year follow-up study. Lancet Diabetes Endocrinol. 7, 684–694 (2019).
Dennis, J. M., Shields, B. M., Henley, W. E., Jones, A. G. & Hattersley, A. T. Disease progression and treatment response in data-driven subgroups of type 2 diabetes compared with models based on simple clinical features: an analysis using clinical trial data. Lancet Diabetes Endocrinol. 7, 442–451 (2019).
Bello-Chavolla, O. Y. et al. Clinical characterization of data-driven diabetes subgroups in Mexicans using a reproducible machine learning approach. BMJ Open Diabetes Res. Care 8, e001550 (2020).
Rasouli, B. et al. Smoking and the risk of LADA: results from a Swedish population-based case-control study. Diabetes Care 39, 794–800 (2016).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Sharp, S. A. et al. Development and standardization of an improved type 1 diabetes genetic risk score for use in newborn screening and incident diagnosis. Diabetes Care 42, 200–207 (2019).
Wood, A. R. et al. A genome-wide association study of IVGTT-based measures of first-phase insulin secretion refines the underlying physiology of type 2 diabetes variants. Diabetes 66, 2296–2309 (2017).
Prokopenko, I. et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 10, e1004235 (2014).
Manning, A. K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat. Genet. 44, 659–669 (2012).
Walford, G. A. et al. Genome-wide association study of the modified Stumvoll Insulin Sensitivity Index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 65, 3200–3211 (2016).
Mahajan, A. et al. Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 50, 559–571 (2018).
Udler, M. S. et al. Type 2 diabetes genetic loci informed by multi-trait associations point to disease mechanisms and subtypes: a soft clustering analysis. PLoS Med. 15, e1002654 (2018).
Cousminer, D. L. et al. First genome-wide association study of latent autoimmune diabetes in adults reveals novel insights linking immune and metabolic diabetes. Diabetes Care 41, 2396–2403 (2018).
Lyssenko, V. et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 117, 2155–2163 (2007).
Spracklen, C. N. et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 582, 240–245 (2020).
Kamat, M. A. et al. PhenoScanner V2: an expanded tool for searching human genotype–phenotype associations. Bioinformatics 35, 4851–4853 (2019).
Staley, J. R. et al. PhenoScanner: a database of human genotype–phenotype associations. Bioinformatics 32, 3207–3209 (2016).
Pirinen, M. et al. Assessing allele-specific expression across multiple tissues from RNA-seq read data. Bioinformatics 31, 2497–2504 (2015).
Tuomi, T. et al. Clinical and genetic characteristics of type 2 diabetes with and without GAD antibodies. Diabetes 48, 150–157 (1999).
Barry, E. et al. Efficacy and effectiveness of screen and treat policies in prevention of type 2 diabetes: systematic review and meta-analysis of screening tests and interventions. BMJ 356, i6538 (2017).
Rosvall, M. et al. Risk factors for the progression of carotid intima-media thickness over a 16-year follow-up period: the Malmö Diet and Cancer Study. Atherosclerosis 239, 615–621 (2015).
Groop, L. et al. Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes 45, 1585–1593 (1996).
Anderson, C. A. et al. Data quality control in genetic case-control association studies. Nat. Protoc. 5, 1564–1573 (2010).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
Nagy, R. et al. Exploration of haplotype research consortium imputation for genome-wide association studies in 20,032 Generation Scotland participants. Genome Med. 9, 23 (2017).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Turner, S. D. qqman: an R package for visualizing GWAS results using Q–Q and manhattan plots. Preprint at bioRxiv https://doi.org/10.1101/005165 (2014).
Acknowledgements
This study was supported by grants from the Swedish Research Council (project grant nos. 2017-02688 and 2020-02191 to E.A.), project grant no. 2015-2558 and infrastructure grant nos. 2010-5983, 2012-5538 and 2014-6395, Linnaeus grant no. 349-2006-237, a strategic research grant no. 2009-1039 (EXODIAB) all to L.G., project grant no. 2018-02635 to O.H. and Indo-Swedish joint network grant no. 2015-06722 to R.B.P.; a Vinnova Swelife grant (no. 2017-05284); and grants from the Academy of Finland (nos. 263401 and 267882 to L.G.), Novo Nordisk Foundation (no. NNF18OC0034408), Scania University Hospital (ALF grant), Diabetes Wellness Sweden (no. 25-420 PG), the Swedish Heart and Lung Foundation (no. 20180640), all to E.A., the Swedish Diabetes Foundation (no. DIA2019-468 to E.A., no. DIA2020-511 to O.H.), the EFSD/Lilly European Diabetes Research Programme to E.A., the Hjelt Foundation and the Albert Påhlsson Research Foundation to E.A. and O.H., the Crafoord Foundation (nos. 20180778, 20190912 and 20200867 to E.A., no. 20200828 to O.H. and no. 20200891 to R.B.P.) and Åke Wiberg Foundation (no. M20-0214) to R.B.P. This project was also financially supported by the Swedish Foundation for Strategic Research (no. IRC15-0067). The genotyping of ANDIS was funded by Pfizer. DIREVA was supported by the Vaasa Hospital District and the Jakobstadsnejden Heart, Folkhalsan Research and Ollqvist Foundations (to T.T. and A.K.). The Botnia Study (L.G., T.T.) has been supported by grants from the Folkhälsan Research and Sigrid Juselius Foundations, Academy of Finland (grants nos. 263401, 267882 and 312063 to L.G., 312072 to T.T. and 317599 to O.P.D.), University of Helsinki, Nordic Center of Excellence in Disease Genetics, European Union (EXGENESIS FP6-2004-005272, MOSAIC FP7-600914), Ollqvist Foundation, Swedish Cultural Foundation in Finland, Finnish Diabetes Research Foundation, Foundation for Life and Health in Finland, Signe and Ane Gyllenberg Foundation, Finnish Medical Society, Paavo Nurmi Foundation, State Research Funding via Helsinki University Hospital and the Perklén, Närpes Health Care and Ahokas Foundations. The study has also been supported by the Ministry of Education in Finland, Municipal Heath Care Center and Hospital in Jakobstad and Health Care Centers in Vaasa, Närpes and Korsholm. The research leading to these results has received funding from the European Research Council under the European Union’s Seventh Framework Programme (no. FP7/2007-2013)/ERC grant agreement no. 269045 and the Innovative Medicines Initiative 2 Joint Undertaking (no. 115881, RHAPSODY). The skillful assistance of the Botnia Study Group is gratefully acknowledged. The genotyping of Botnia was funded by Regeneron Pharmaceuticals. The study sponsors had no role in the analysis and interpretation of the data, writing of the manuscript or decision to submit the manuscript for publication. E.A. had full access to all the data in the study and had the final responsibility for the decision to submit for publication. We thank all patients and healthcare providers for their support and willingness to participate. We also thank J. Hultman, M. Borell, J. Kravic, G. Gremsperger, M. Sterner, M. Neptin and U. Blom-Nilsson for excellent technical and administrative support, Region Skåne (Scania County) and the ANDIS steering committee for their support.
Author information
Authors and Affiliations
Consortia
Contributions
D.M.A., O.P.D., R.B.P., J.C.F., M.I.M., T.T., L.G. and E.A. contributed to the conception of the work. A.K., J.B., R.G.C., O.M., S.C., O.H., T.T., L.G. and E.A. contributed to data collection. D.M.A., O.P.D., R.H., M.T., M.Å., A.M., M.S.U. and E.A. contributed to the data analysis. D.M.A., O.P.D., T.T., L.G. and E.A. drafted the manuscript. All authors contributed to the interpretation of the data and critical revision of the manuscript. All authors gave final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The views expressed in this article are those of the authors and not necessarily those of the National Health Service, National Institute for Health Research or Department of Health. M.I.M. has served on advisory panels for Pfizer, Novo Nordisk and Zoe Global, has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly and research funding from AbbVie, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. As of January 2020, A.M. is an employee of Genentech and a holder of Roche stock. J.B. is an employee and shareholder of Pfizer. J.C.F. has received consulting honoraria from Goldfinch Bio and speaking honoraria from Novo Nordisk. The other authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Stephen Rich, Constantin Polychronakos, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Association of GRS of diabetic complications with diabetes subtypes.
a-e, Association of GRS for chronic kidney disease (CKD) (a), estimated glomerular filtration rate (eGFR) (b), urine albumin creatinine ratio (UACR) (c), diabetic retinopathy (DR) (d) and cardiovascular disease (CVD) (e). Each subtype was compared to diabetes-free controls using logistic regression adjusted for sex and PCs. ANDIS (nSAID = 452, nSIDD = 1,193, nSIRD = 1,129, nMOD = 1,374, nMARD = 2,861, nT2D = 7,676, ncontrols = 2,744), and DIREVA (nSAID = 327, nSIDD = 394, nSIRD = 453, nMOD = 596, nMARD = 1,178, nT2D = 2,621, ncontrols = 1,683). In ANDIS, eGFR-GRS, UACR-GRS and CVD-GRS showed no association with any subtype. CKD-GRS was associated with SIRD with nominal significance in ANDIS (P = 0.047) but not in DIREVA (P = 0.38). DR was associated with SIRD (P = 0.016) and MOD (P = 0.015) with nominal significance but not in DIREVA (P = 0.37 and P = 0.36). None of the GRS were significant after adjustment for multiple comparisons (n = 5 tests). Data are presented as odds ratio and 95% confidence interval per s.d. increase of GRS. Exact numbers and statistics are in Supplementary Table 13. P-values are two-sided and not adjusted for multiple comparisons.
Extended Data Fig. 2 Manhattan and QQ plots of GWAS in ANDIS-MDC.
a-e, GWAS of each subtype in ANDIS using diabetes-free controls (n = 2,744) from the MDC study: SAID (n = 452) (a), SIDD (n = 1,193) (b), SIRD (n = 1,130) (c), MOD (n = 1,374) (d), MARD (n = 2,861) (e). Variants in the HLA region were associated with SAID (rs9273368 OR = 2.89 [2.31-3.10], P = 6.5 ×10−40). The rs7903146TCF7L2 locus was associated with SIDD (OR = 1.56 [1.39-1.75], P = 8.6 ×10−15), MOD (OR = 1.40 [1.26-1.56], P = 3.1 ×10−10) and MARD (1.42 [1.30-1.54], P = 6.1 ×10−16). rs10824307 near the LRMDA gene was associated with MOD (OR = 1.35 [1.22-1.49], P = 1.3 ×10−9). The red line indicates the genome-wide significance threshold (P < 5 ×10−8) and the blue line suggestive association (P < 10−5). Statistics were calculated using the additive SNPtest score method adjusting for sex and 4 PCs. P-values are two-sided and not adjusted for multiple comparisons.
Supplementary information
Supplementary Table 1
Supplementary Tables 1–16
Rights and permissions
About this article
Cite this article
Mansour Aly, D., Dwivedi, O.P., Prasad, R.B. et al. Genome-wide association analyses highlight etiological differences underlying newly defined subtypes of diabetes. Nat Genet 53, 1534–1542 (2021). https://doi.org/10.1038/s41588-021-00948-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-021-00948-2
This article is cited by
-
Randomized open-label trial of semaglutide and dapagliflozin in patients with type 2 diabetes of different pathophysiology
Nature Metabolism (2024)
-
A second step towards precision medicine in diabetes
Nature Metabolism (2024)
-
Metabolic profiling of smoking, associations with type 2 diabetes and interaction with genetic susceptibility
European Journal of Epidemiology (2024)
-
Association between a polygenic lipodystrophy genetic risk score and diabetes risk in the high prevalence Maltese population
Acta Diabetologica (2024)
-
Age-dependent topic modeling of comorbidities in UK Biobank identifies disease subtypes with differential genetic risk
Nature Genetics (2023)