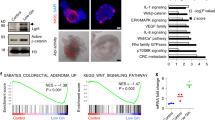
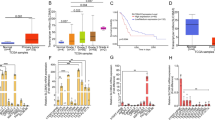
Abstract
Oncogenic KRAS mutations and inactivation of the APC tumor suppressor co-occur in colorectal cancer (CRC). Despite efforts to target mutant KRAS directly, most therapeutic approaches focus on downstream pathways, albeit with limited efficacy. Moreover, mutant KRAS alters the basal metabolism of cancer cells, increasing glutamine utilization to support proliferation. We show that concomitant mutation of Apc and Kras in the mouse intestinal epithelium profoundly rewires metabolism, increasing glutamine consumption. Furthermore, SLC7A5, a glutamine antiporter, is critical for colorectal tumorigenesis in models of both early- and late-stage metastatic disease. Mechanistically, SLC7A5 maintains intracellular amino acid levels following KRAS activation through transcriptional and metabolic reprogramming. This supports the increased demand for bulk protein synthesis that underpins the enhanced proliferation of KRAS-mutant cells. Moreover, targeting protein synthesis, via inhibition of the mTORC1 regulator, together with Slc7a5 deletion abrogates the growth of established Kras-mutant tumors. Together, these data suggest SLC7A5 as an attractive target for therapy-resistant KRAS-mutant CRC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-seq data that support the findings of this are available at GEO (accession nos. GSE160478 and GSE160479). Human data analyzed in this study are publicly available through TCGA at https://gdc.cancer.gov/about-data/publications/pancanatlas. Source data are provided with this paper.
References
Muzny, D. M. et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Faller, W. J. et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500 (2015).
Maddocks, O. D. K. et al. Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376 (2017).
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2013).
Hung, K. E. et al. Development of a mouse model for sporadic and metastatic colon tumors and its use in assessing drug treatment. Proc. Natl Acad. Sci. USA 107, 1565–1570 (2010).
Haigis, K. M. et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat. Genet. 40, 600–608 (2008).
Ng, K. et al. Phase II study of everolimus in patients with metastatic colorectal adenocarcinoma previously treated with bevacizumab-, fluoropyrimidine-, oxaliplatin-, and irinotecan-based regimens. Clin. Cancer Res. 19, 3987–3995 (2013).
Spindler, K.-L. G. et al. Phase II trial of temsirolimus alone and in combination with irinotecan for KRAS mutant metastatic colorectal cancer: outcome and results of KRAS mutational analysis in plasma. Acta Oncol. 52, 963–970 (2013).
Gimple, R. C. & Wang, X. RAS: striking at the core of the oncogenic circuitry. Front. Oncol. 9, 965 (2019).
White, E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 27, 2065–2071 (2013).
Son, J. et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature 496, 101–105 (2013).
Mayers, J. R. & Vander Heiden, M. G. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem. Sci. 40, 130–140 (2015).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21, 1350–1356 (2015).
Davidson, S. M. et al. Environment impacts the metabolic dependencies of Ras-driven non-small cell lung cancer. Cell Metab. 23, 517–528 (2016).
Biancur, D. E. et al. Compensatory metabolic networks in pancreatic cancers upon perturbation of glutamine metabolism. Nat. Commun. 8, 15965 (2017).
Mayers, J. R. et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 (2016).
McBrayer, S. K. et al. Transaminase inhibition by 2-hydroxyglutarate impairs glutamate biosynthesis and redox homeostasis in glioma. Cell 175, 101–116 (2018).
César-Razquin, A. et al. A call for systematic research on solute carriers. Cell 162, 478–487 (2015).
Nicklin, P. et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 136, 521–534 (2009).
Saito, Y. et al. LLGL2 rescues nutrient stress by promoting leucine uptake in ER+ breast cancer. Nature 569, 275–279 (2019).
Collins, M. A. et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 122, 639–653 (2012).
Elorza, A. et al. HIF2α acts as an mTORC1 activator through the amino acid carrier SLC7A5. Mol. Cell 48, 681–691 (2012).
Grzes, K. M. et al. Control of amino acid transport coordinates metabolic reprogramming in T-cell malignancy. Leukemia 31, 2771–2779 (2017).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Di Nicolantonio, F. et al. Deregulation of the PI3K and KRAS signaling pathways in human cancer cells determines their response to everolimus. J. Clin. Invest. 120, 2858–2866 (2010).
Buck, E. et al. Rapamycin synergizes with the epidermal growth factor receptor inhibitor erlotinib in non-small-cell lung, pancreatic, colon, and breast tumors. Mol. Cancer Ther. 5, 2676–2684 (2006).
Skrzypczak, M. et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS ONE 5, e13091 (2010).
Dong, H. K. et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int. J. Cancer 121, 2005–2012 (2007).
Kaiser, S. et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 8, R131 (2007).
Marisa, L. et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 10, e1001453 (2013).
Jackstadt, R. et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell 36, 319–336 (2019).
Gaglio, D. et al. Oncogenic K‐Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 7, 523 (2011).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017).
Hosios, A. M. et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell 36, 540–549 (2016).
Commisso, C. et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Shibata, H. et al. Rapid colorectal adenoma formation initiated by conditional targeting of the APC gene. Science 278, 120–133 (1997).
Jackson, E. L. et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15, 3243–3248 (2001).
Sinclair, L. V. et al. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 14, 500–508 (2013).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Jonkers, J. et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001).
Murtaugh, L. C., Stanger, B. Z., Kwan, K. M. & Melton, D. A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl Acad. Sci. USA 100, 14920–14925 (2003).
Finlay, M. R. V. et al. Discovery of a thiadiazole–pyridazine-based allosteric glutaminase 1 inhibitor series that demonstrates oral bioavailability and activity in tumor xenograft models. J. Med. Chem. 62, 6540–6560 (2019).
Dannhorn, A. et al. Universal sample preparation unlocking multimodal molecular tissue imaging. Anal. Chem. 92, 11080–11088 (2020).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Race, A. M., Styles, I. B. & Bunch, J. Inclusive sharing of mass spectrometry imaging data requires a converter for all. J. Proteomics 75, 5111–5112 (2012).
Race, A. M. et al. SpectralAnalysis: software for the masses. Anal. Chem. 88, 9451–9458 (2016).
Dexter, A., Race, A. M., Styles, I. B. & Bunch, J. Testing for multivariate normality in mass spectrometry imaging data: a robust statistical approach for clustering evaluation and the generation of synthetic mass spectrometry imaging data sets. Anal. Chem. 88, 10893–10899 (2016).
Sakamaki, J. I. et al. Bromodomain protein BRD4 Is a transcriptional repressor of autophagy and lysosomal function. Mol. Cell 66, 517–532 (2017).
Liu, R. et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 43, e97 (2015).
Charif, D. & Lobry, J. R. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis 207–232 (Springer, 2007).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128 (2013).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Lewis, D. Y. et al. [18F]fluoroethyltyrosine-induced Cerenkov luminescence improves image-guided surgical resection of glioma. Theranostics 8, 3991–4002 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We are grateful to the CRUK Beatson Institute Central Services, Histology Service, the Transgenic Technology Laboratory and the Biological Services Unit for technical support and all members of the Sansom lab for discussion of the data and manuscript. O.J.S. was supported by grants from CRUK (A21139, A12481 and A17196) and the ERC (311301). A.K.N, G.H., R.T.S., C.J.N., T.M., A.M.R., A.D., Z.T., R.J.A.G. and J.B. were supported by the CRUK Rosetta Grand Challenge grant (A25045 to J.B. and O.J.S.). F.C., R.A.R., D. Sumpton, C.N., G.M.M., E.G., S. Tardito and A.D.C. were supported by the CRUK Beatson Institute core grant (A17196 to O.J.S.). S.K.F. was supported by Pancreatic Cancer UK Future Leaders Academy (to O.J.S.). N.V. was supported by the Wellcome Trust (201487 to O.J.S.). D.M.G. was supported by an ERC grant (311301 to O.J.S.). J.R.P.K. was supported by a CRUK Programme Award (A24388 to M.B. and O.J.S.). R.J. was supported by a Marie Skłodowska-Curie Actions postdoctoral fellowship (ERC 659666). J.D.G.L. was supported by an MRC Clinical Research Training Fellowship (MR/N021800/1). K.G. was supported by the CRUK Grand Challenge Specificancer Grand Challenge Consortium (A29055 to O.J.S.). Holly Hall. was supported by BB/N017005/1. M.B. was supported by the CRUK Beatson Institute core grant 29252. S.B.M. and P.D.D. were supported by a CRUK Early Detection award (C50686/A29834). GLS inhibitor (described in ref. 44) was provided by AstraZeneca and CRUK Therapeutic Discovery Laboratories. We thank M. Pasca di Magliano (University of Michigan) for providing us with the doxycycline-inducible KrasG12D mouse. We thank A. Dannhorn (AstraZeneca) for establishment of the MSI sample preparation method and A. Dannhorn and N. Strittmatter (AstraZeneca) for DESI optimization. We thank the members of the CRUK Rosetta Grand Challenge Consortium for participating in this study.
Author information
Authors and Affiliations
Consortia
Contributions
A.K.N., F.C., A.D.C. and O.J.S. designed the research. A.K.N., F.C., S.K.F., D.M.G., N.V., J.R.P.K., R.A.R., D. Sumpton and G.M.M. performed the experiments and analyzed data. A.K.N., E.R.J., A.M., G. Malviya and D.Y.L. performed PET–MRI and analyzed the results. D. Solovyev and G.B. radiosynthesized 18F-FET. A.K.N., F.C., D. Sumpton, G.R.-B., G.M.M., E.G. and S. Tardito performed and analyzed metabolomic experiments. A.K.N., Holly Hall, K.G., W.C., A.H. and M.B. performed transcriptomics and analyzed transcriptomic data. A.K.N., K.G., S.B.M. and P.D.D. analyzed human data. J.D.G.L. helped derive iKras organoid lines. R.J. and J.D.G.L. helped analyze KPN mice. A.K.N. and C.N. performed IHC. G.H., R.T.S., C.J.N., T.M., A.M.R., A.D., Z.T., R.J.A.G. and J.B. performed and analyzed MSI data. S.E.C. provided the GLS inhibitor. D. Strathdee rederived Slc7a5fl/fl mice. S.T.B. and Z.T. provided expertise on MSI and critical feedback. A.K.N., A.D.C. and O.J.S. wrote the paper. All authors read the manuscript and provided critical comments.
Corresponding author
Ethics declarations
Competing interests
S.T.B. is an employee and shareholder of AstraZeneca. The other authors declare no competing interests.
Additional information
Peer review information Nature Genetics thanks Thales Papagiannakopoulos, Louis Vermeulen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Kras activation drives metabolic reprogramming and confers resistance to GLS inhibition.
a, Normalized enrichment scores (NES) of metabolic signature gene sets from ‘Hallmark’ gene set collection in Apcfl/fl KrasG12D/+ mouse intestine. Gene sets with nominal P values after multiple hypotheses correction and FDR, q < 0.05 indicated for each gene set. b, Diagram of glycolysis and glutaminolysis pathway intermediates. 13C-glucose (13C6)-labelled carbons (red); 13C-Glutamine (13C5)-labelled carbons (blue). c, Schematic illustrating role of GLS in glutaminolysis. d, Glutamine hydrolysis in Apcfl/fl KrasG12D/+ organoids after 24 h treatment with GLS inhibitor (GLSi; 1 µM), CB839 (1 µM), BPTES (20 µM), or DMSO (vehicle) followed by 13C5-Glutamine–tracing using LC-MS. Plots show mean percentage of 13C5-glutamine-derived 13C5-glutamate (peak area/µg of protein) ± s.e.m. Each treatment performed as an independent experiment. *P =0.05, one-way Mann–Whitney U test. e, Relative viability (mean ± s.e.m.) of Apcfl/fl KrasG12D/+ organoids treated as in d for 72 h. f, H&E and BrdU staining showing resistance to GLSi treatment for 72 h in the small intestine (SI) and colon of Apcfl/fl KrasG12D/+ mice (representative of 4 biologically independent mice). Treatment began 24 h after induction. Scale bars, 100 µm. g, Number of BrdU-positive cells per half-crypt (mean ± s.e.m.) in SI and colon from (f). n=4 biological replicates per group; 25 half-crypts were scored per mouse. h, Schematic illustrating leucine or valine transamination by BCAT and nitrogen (orange ovals) fate after transamination. i, Plots show mean percentage of 15N-leucine-derived or 15N-valine-derived glutamate (peak area/µg of protein) ± s.e.m. j, Mass spectrometry imaging (MSI) of 13C5-glutamine derived TCA cycle intermediates in APC and APC KRAS small intestinal tissue. Plots show ion abundance of isotope-labelled glutamine-derived intermediates ± s.e.m. n = 4 biological replicates per group. P using two-way ANOVA, with Tukey’s multiple comparison test. In d, e, i n = 3 biological replicates in technical triplicates each per group.
Extended Data Fig. 2 KrasG12D mediates Slc7a5 expression and metabolic reprogramming.
a, RNA in situ hybridization (ISH) images showing Slc7a5 and Slc1a5 expression. Dashed boxes highlight areas shown in high magnification. Scale bars, 200 µm. b, qRT-PCR analysis of indicated genes in mouse intestinal tissues. Transcript levels are shown as delta–delta threshold cycle (ΔΔCT) relative to Gapdh. c, Top, Slc7a5 immunoblotting in intestinal tissue lysates. Representative of three independent experiments. ß-actin as loading control. Bottom, bar graph showing Slc7a5 protein level quantification, normalized to ß-actin. d, Left, ISH images showing Slc7a5 expression in Apcfl/+ and Apcfl/+ KrasG12D/+ tumors. Scale bars, 100 µm. Right, bar graph showing quantification of Slc7a5 positivity calculated as mean percentage (Apcfl/+, n = 5; Apcfl/+ KrasG12D/+, n = 4). e, Slc7a5 substrates in small intestinal tissues as measured by MSI. Boxes depict interquartile range, central line indicates median and whiskers indicate minimum/maximum values (WT, Apcfl/fl, Apcfl/fl KrasG12D/+, n = 5 and KrasG12D/+, n = 4). f, Schematic of genetic crossing to generate Apcfl/+ iKrasG12D mice. g, Brightfield images of Apc iKrasG12D organoid lines in presence or absence of Doxycycline for 48 hours. Representative of four individual wells from three independent lines. Scale bar 100 μm. h, Relative expression of metabolic genes in Apc iKrasG12D organoids from RNAseq reads normalized to the presence of Dox. i, Heatmap shows relative abundance of metabolites (peak area/µg of protein) upon doxycycline withdrawal in Apc iKrasG12D organoid lines using LC-MS. Data represent mean (n = 3 biological replicates in technical triplicates each per group). In a, d images representative of 4 mice per genotype. b, c, h n = 3 per genotype. b, c, d, h data are mean ± s.e.m. P using two-way ANOVA, with Tukey’s test (b), Fisher’s test (h), one-way Mann–Whitney U test (c, d) and one-way ANOVA, with Tukey’s test (e).
Extended Data Fig. 3 Deletion of Slc7a5 does not affect intestinal morphology, KrasG12D mediated intestinal phenotypes or Wnt activation in Apcfl/fl KrasG12D/+ mice.
a, Schematic of the experimental approach to investigate intestinal homeostasis. b, H&E, BrdU IHC, and ISH for Wnt-target genes Ascl2, Lgr5 and Olfm4 in the small intestine, after Slc7a5 deletion, compared to Slc7a5+/+. Representative of four biologically independent mice for each genotype. Scale bar, 100 µm. c, Schematic of the experimental approach to investigate the phenotypes associated with prolonged KRAS activation. d, Length of small intestine (SI) and colon of KrasG12D/+ Slc7a5+/+ and KrasG12D/+ Slc7a5fl/fl mice sampled 30 days post Cre-induction. Data are mean ± s.e.m. (KrasG12D/+ Slc7a5+/+, n = 5; KrasG12D/+ Slc7a5fl/fl, n = 4). e, Number of crypts per circumference in the small intestine of KrasG12D/+ Slc7a5+/+ and KrasG12D/+ Slc7a5fl/fl mice, sampled 30 days post Cre-induction. At least 10 circumferences per mouse were scored. Data are mean ± s.e.m. (KrasG12D/+ Slc7a5+/+, n = 3; KrasG12D/+ Slc7a5fl/fl, n = 4). f, Representative IHC for β-catenin, showing no significant differences between Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D Slc7a5fl/fl mice (n = 3 biological replicates). Red line indicates the small intestinal crypt height. Scale bar, 200 µm.
Extended Data Fig. 4 Characterization of Slc7a5-deficient Apcfl/+ KrasG12D/+ tumors.
a, Macroscopic size distribution analysis of intestinal tumors (in millimetres) at endpoint (Apcfl/+ KrasG12D/+ Slc7a5+/+ and Apcfl/+ KrasG12D/+ Slc7a5fl/fl, n = 15 per genotype). b, Number of tumors quantified in the intestine at 60 days post Cre-induction (Apcfl/+ KrasG12D/+ Slc7a5+/+ and Apcfl/+ KrasG12D/+ Slc7a5fl/fl, n = 3 per genotype). Data are mean ± S.E.M. P = 0.05, one-way Mann–Whitney U test. c, Left, distribution of bottom-up (crypt-derived) and top-down (villus-derived) intestinal tumors at endpoint (Apcfl/+ KrasG12D/+ Slc7a5+/+, n = 9; Apcfl/+ KrasG12D/+ Slc7a5fl/fl, n = 15). Right, H&E showing a top-down tumor from Apcfl/fl KrasG12D/+ Slc7a5fl/fl mice at endpoint. Representative of eight biologically independent Apcfl/+ KrasG12D/+ Slc7a5fl/fl mice. Scale bar, 100 μm. d, Representative ISH for Slc7a5 and IHC for nuclear β-catenin in Apcfl/+ KrasG12D/+ Slc7a5+/+ and Apcfl/+ KrasG12D/+ Slc7a5fl/fl tumors. Representative of four biologically independent mice for each genotype. Scale bars, 100 μm. e, Heatmap showing relative abundance of metabolites (metabolite peak area/µg of protein) in small intestinal and colon tissues from Apcfl/fl KrasG12D/+ Slc7a5+/+ (n = 5) and Apcfl/fl KrasG12D/+ Slc7a5fl/fl (n = 4) mice as measured by LC-MS.
Extended Data Fig. 5 Mass spectrometry imaging reveals metabolic alterations following Slc7a5 deletion.
a, Schematic of workflow for LC-MS and mass spectrometry imaging (MSI) of intestinal tissue. b, H&E and ion intensities of glutamine, glutamate and alanine in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl small intestinal tissue, using MSI. Representative ion images from three biologically independent mice for each genotype. c, Table showing ion intensities from b. d, Abundance of amino acids (metabolite peak area/µg of protein) in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl small intestinal tissues using LC-MS. Heatmap shows mean relative abundance (n = 4 biological replicates per group). Asterisks denote P, 0.001 (Met), 0.004 (His), 0.01 (Gly), 0.02 (Pro), 0.02 (Asp), 0.003 (Lys), 0.0001 (Ser), 0.0005 (Thr), 0.000006 (Arg), 0.002 (Gln), using two-way ANOVA with Fisher’s test. e, H&E and autoradiogram of O-(2-18F-fluoroethyl)-L-tyrosine (18F-FET) uptake and distribution in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl mice. Representative images from four biologically independent mice for each genotype. f, H&E and ion intensities of leucine, 13C6-leucine, KIC and KIC M+6 in WT, KrasG12D/+, Apcfl/fl, Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl small intestinal tissue, generated using MSI. Representative ion images from four individual mice for each genotype. g, Ratio of ion abundances of KIC M+6 and 13C6-leucine. Plot shows mean ratios ± s.e.m. h, Ion abundance of citrate M+2 (detected with MSI) and peak area of acetyl-CoA M+2 (detected with LC-MS). Plots show their respective mean ± s.e.m. In g, h WT, n =5, Apcfl/fl, Apcfl/fl KrasG12D/+, KrasG12D/+, and Apcfl/fl KrasG12D/+ Slc7a5fl//fl, n = 4; per group. P using one-way ANOVA followed by Tukey’s test. In b, f selected ion images representing the accurate mass distribution of indicated metabolites are shown. Ion intensities were scaled individually for each metabolite and m/z ratios indicated.
Extended Data Fig. 6 Loss of Slc7a5 in Lgr5-positive cells does not alter Apc tumorigenesis or mTORC1 signaling.
a, Kaplan–Meier survival curve of Lgr5CreER Apcfl/fl Slc7a5+/+ and Lgr5CreER Apcfl/fl Slc7a5fl/fl mice aged until clinical endpoint (Lgr5CreER Apcfl/fl Slc7a5+/+, n = 9; Lgr5CreER Apcfl/fl Slc7a5fl/fl, n = 15). P = 0.1860, log-rank (Mantel–Cox) test. b, Boxplot showing total number of tumors from Lgr5CreER Apcfl/fl and Lgr5CreER Apcfl/fl Slc7a5fl/fl mice aged until clinical endpoint. Lgr5CreER Apcfl/fl, n = 9; Lgr5CreER Apcfl/fl Slc7a5fl/fl, n = 15. Box depicts the interquartile range, central line indicates the median and whiskers indicate minimum/maximum values. c, Representative H&E, BrdU, pS6 and peEF2 staining of tumors from Lgr5CreER Apcfl/fl Slc7a5+/+ and Lgr5CreER Apcfl/fl Slc7a5fl/fl mice (representative of three biologically independent mice for each genotype). Scale bar, 100 μm. d, Representative pmTOR, peEF2 and p4EBP1 staining in Apcfl/+ KrasG12D/+ Slc7a5+/+ and Apcfl/+ KrasG12D/+ Slc7a5fl/fl tumors (representative of three biologically independent mice for each genotype). Dashed boxes highlight selected areas shown in high magnification. Scale bar, 100 μm.
Extended Data Fig. 7 Interplay between SLC7A5, essential amino acids, and protein synthesis.
a, qRT-PCR analysis of indicated genes in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal organoids. Transcript levels were normalized to Gapdh (n = 3 per group). Data are mean ± s.e.m. P = 2 x 10-6 using two-way ANOVA with Sidak’s multiple comparisons test. b, Immunoblotting of Slc7a5 in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal organoids. Each lane corresponds to organoids from a biologically independent mouse. Representative of two independent experiments. HSP90 was used as a loading control. c, Measurement of 35S-methionine uptake in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal crypt cultures. Uptake of 35S was normalized to total protein. Data are mean ± s.e.m. (n = 3 biological replicates in technical duplicates per group). P = 0.5, one-way Mann–Whitney U test. d, Representative ISH for pS6 and p4EBP1 in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl mice (n = 3 biological replicates). Scale bar, 100 μm. e, Ribosome run-off rate measured in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal organoids. The run-off rate was measured by addition of translation elongation inhibitor Harringtonine for 0 or 180 s, and increase in sub-polysomes relative to polysomes was calculated. The shift between two timepoints’ S:P ratio, represents elongation speed. Data are mean ± s.e.m. (n = 3 biological replicates per group). P = 0.42 using one-way Mann–Whitney U test. f, Representative polysome profiles of intestinal epithelial cells showing RNA distribution in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl mice (n = 3 biological replicates per group). g, Quantification of protein synthesis 35S- methionine in Apcfl/fl KrasG12D/+ Slc7a5+/+ and Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal organoids 16 h after depletion of leucine/isoleucine. Data are mean ± s.e.m. (n = 3 biological replicates per group). *P = 0.02 using one-way Mann–Whitney U test.
Extended Data Fig. 8 SLC7A5 deficiency leads to transcriptional reprogramming towards survival.
a, Enrichment scores of gene sets from KEGG collection in Apcfl/fl KrasG12D/+ Slc7a5fl/fl mouse intestine. Gene sets up- and downregulated are highlighted in red and green, respectively. b, Bar graph showing top 20 GO biological processes terms enriched in Apcfl/fl KrasG12D/+ Slc7a5fl/fl mice. c, qRT-PCR analysis of endoplasmic reticulum stress/unfolded protein response genes in Apcfl/fl KrasG12D/+ Slc7a5fl/fl intestinal tissues. Transcript levels were normalized to Gapdh and relative to Apcfl/fl KrasG12D/+ Slc7a5+/+ controls (n = 3 per group). Data are mean ± s.e.m, asterisks denote P, 0.03 (Atf4), 1.1 x 10-15 (Trib3), 0.008 (Chac1), < 10-15 (Slc7a3), 0.08 (Asns), 0.01 (Psph), 2.3 x 10-9 (Slc7a11), from two-way ANOVA with Fisher’s test. d, Immunostaining of LC3 puncta in Apcfl/+ KrasG12D/+ Slc7a5+/+ and Apcfl/+ KrasG12D/+ Slc7a5fl/fl intestinal tumors. Plot showing mean LC3 puncta/cell in tumors. Box depicts interquartile range, central line indicates median and whiskers indicate minimum/maximum values. P using one-tailed Mann–Whitney U test. Scale bar, 100 μm. e, Boxplots showing length distributions of 5’ UTR, coding, and 3’ UTR in downregulated (red) and upregulated (blue) genes (dots) in Apcfl/fl KrasG12D/+ mice treated with rapamycin. y-axis shows length of each region on log10 scale. Central line indicates median, lower line is first quantile (Q1) and upper line is third quantile (Q3). Upper whisker extends from Q3 to 1.5 times inter-quartile range (IQR) and lower whisker from Q1 to smallest value at most 1.5 times IQR. P using, non-parametric post-hoc Dunn’s test with Bonferroni correction following one-way ANOVA. f, Bar graph showing top 20 GO biological processes terms enriched among in rapamycin-treated Apcfl/fl KrasG12D/+ mice relative to vehicle controls. In b, f GO analysis was performed with Enrichr using standard settings; P value following Fisher exact Test is plotted for each term. In d, e n = 4 per group.
Extended Data Fig. 9 Rapamycin reduces Slc7a5-deficient cell proliferation and tumor formation.
a, Kaplan–Meier survival curves of Apcfl/+ KrasG12D/+ Slc7a5+/+, untreated Apcfl/+ KrasG12D/+ Slc7a5fl/fl, and rapamycin-treated Apcfl/+ KrasG12D/+ Slc7a5fl/fl mice recovering from clinical symptoms of intestinal tumor burden. Rapamycin treatment (10 mg/kg) of Apcfl/+ KrasG12D/+ Slc7a5fl/fl mice was initiated when mice showed signs of intestinal disease (approximately 120 days post-induction), and lasted until mice were censored due to health issues unrelated to intestinal tumors. (Apcfl/+ KrasG12D/+ Slc7a5+/+ and untreated Apcfl/+ KrasG12D/+ Slc7a5fl/fl, n = 15 mice per group; Apcfl/+ KrasG12D/+ Slc7a5fl/fl + Rapamycin, n = 6 mice). b, Top, schematic representation of rapamycin and everolimus treatment strategy. Treatment (10 mg/kg) was initiated when mice showed signs of intestinal disease and continued for 5 days thereafter. Lower left, BrdU IHC showing that short-term rapamycin or everolimus treatment inhibits tumor proliferation in Apcfl/+ KrasG12D/+ Slc7a5fl/fl mice but not in SLC7A5-proficient Apcfl/+ KrasG12D/+ counterparts. Lower right, bar graphs showing mean percentage of BrdU-positive cells in intestinal tumors from each genotype (Apcfl/+ KrasG12D/+ Slc7a5+/+, n = 4; Apcfl/+ KrasG12D/+ Slc7a5fl/fl, n = 5 mice per group). Data are mean ± s.e.m. P = 0.007 (Rapamycin), P = 0.004 (Everolimus) using one-tailed Mann–Whitney U test. Scale bar, 100 μm. c, Graphical model of SLC7A5-mTOR-protein synthesis axis in intestinal tumors. Mechanistically, oncogenic KRAS activates a transcriptional programme that augments metabolism. A feature of this metabolic programme includes induction of Slc7a5 expression and influx of amino acids, leading to increased mTORC1 signalling and protein synthesis to meet proliferative demand. Loss of SLC7A5 alters intracellular amino acid homeostasis, and reduces protein synthesis and mTORC1-S6K signalling. While KRAS-mutant cells are resistant to rapamycin, loss of SLC7A5 sensitizes these cells to combinatorial treatment with mTORC1 inhibitors.
Extended Data Fig. 10 Slc7a5 deletion compromises mutant Kras-driven carcinoma formation.
a, Expression of SLC7A5 in patients with colorectal adenocarcinoma versus normal colon from three independent studies (Skrzyczak et al., n = 81 and n = 24; Ki et al., n = 50 and n = 28; Kaiser et al., n = 41 and n = 05, respectively). ****P < 0.0001, unpaired t-test, two-tailed. b, Recurrence free survival (RFS) of CRC patients, stratified using the KRAS mutation status and SLC7A5 expression. Red and blue lines show patients stratified into high SLC7A5 expression (quartile; > Q3) and low SLC7A5 expression (quartile; < Q1), respectively. c, Schematic representing the experimental design to generate KPN SLC7A5 mice. Cre, Cre-recombinase; ER, oestrogen receptor; lox P, Cre-Lox recombination site; IRES, internal ribosome entry site; N1icd, Notch 1 intracellular domain; nEGFP, nuclear-localized enhanced green fluorescent protein; Rosa26, Rosa26 knock-in locus. d, ISH images showing Slc7a5 expression in KPN primary tumor (top) and liver metastasis (bottom). Right images show high magnification of the dashed box in left panels. Dashed line indicates border between tumor (T) and normal epithelium (N) (representative of five biologically independent mice for each genotype). Scale bars, 100 µm. e, Kaplan–Meier survival curves of KrasG12D/+ Trp53fl/fl Slc7a5+/+ and KrasG12D/+ Trp53fl/fl Slc7a5fl/fl mice aged until clinical endpoint (KrasG12D/+ Trp53fl/fl Slc7a5+/+; n=7; KrasG12D/+ Trp53fl/fl Slc7a5fl/fl, n=11). Tick marks indicate censored mice sampled unrelated to intestinal tumor burden. ****P = 2.1 x 10-5, log-rank (Mantel–Cox) test. f, RFS of CRC patients (TCGA) with consensus molecular subtype 4 (CMS4), stratified by low (bottom quartile) or high (top quartile) SLC7A5 expression. Log-rank test P = 0.0219.
Supplementary information
Supplementary Table 1
Primer sequences
Source data
Source Data Extended Data Fig. 2
Unprocessed western blots for Extended Data Fig. 2c
Source Data Extended Data Fig. 7
Unprocessed western blots for Extended Data Fig. 7b
Rights and permissions
About this article
Cite this article
Najumudeen, A.K., Ceteci, F., Fey, S.K. et al. The amino acid transporter SLC7A5 is required for efficient growth of KRAS-mutant colorectal cancer. Nat Genet 53, 16–26 (2021). https://doi.org/10.1038/s41588-020-00753-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-020-00753-3
This article is cited by
-
Fat mass and obesity-associated protein (FTO) mediated m6A modification of circFAM192A promoted gastric cancer proliferation by suppressing SLC7A5 decay
Molecular Biomedicine (2024)
-
Glutamine addiction in tumor cell: oncogene regulation and clinical treatment
Cell Communication and Signaling (2024)
-
A glutamine tug-of-war between cancer and immune cells: recent advances in unraveling the ongoing battle
Journal of Experimental & Clinical Cancer Research (2024)
-
Prediction of metabolites associated with somatic mutations in cancers by using genome-scale metabolic models and mutation data
Genome Biology (2024)
-
Cancer-associated fibroblasts-derived exosomal METTL3 promotes the proliferation, invasion, stemness and glutaminolysis in non-small cell lung cancer cells by eliciting SLC7A5 m6A modification
Human Cell (2024)