Abstract
Multi-tyrosine kinase inhibitors (MTKIs) have thus far had limited success in the treatment of castration-resistant prostate cancer (CRPC). Here, we report a phase I–cleared orally bioavailable MTKI, ESK981, with a novel autophagy inhibitory property that decreased tumor growth in diverse preclinical models of CRPC. The antitumor activity of ESK981 was maximized in immunocompetent tumor environments where it upregulated CXCL10 expression through the interferon-γ pathway and promoted functional T cell infiltration, which resulted in enhanced therapeutic response to immune checkpoint blockade. Mechanistically, we identify the lipid kinase PIKfyve as the direct target of ESK981. PIKfyve knockdown recapitulated ESK981’s antitumor activity and enhanced the therapeutic benefit of immune checkpoint blockade. Our study reveals that targeting PIKfyve via ESK981 turns tumors from cold into hot through inhibition of autophagy, which may prime the tumor immune microenvironment in patients with advanced prostate cancer and be an effective treatment strategy alone or in combination with immunotherapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Raw RNA-seq data have been deposited at the NCBI Gene Expression Omnibus (GSE174644). Further information and requests for resources and reagents should be directed to the corresponding author. All requests for raw and analyzed data and materials will be reviewed promptly by the corresponding author to verify whether the request is subject to any intellectual property or confidentiality obligations. Any data and materials that can be shared will be released via a material transfer agreement. Source data are provided with this paper.
References
Ferraldeschi, R., Welti, J., Luo, J., Attard, G. & de Bono, J. S. Targeting the androgen receptor pathway in castration-resistant prostate cancer: progresses and prospects. Oncogene 34, 1745–1757 (2015).
Scher, H. I. et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N. Engl. J. Med. 367, 1187–1197 (2012).
De Bono, J. S. et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 364, 1995–2005 (2011).
Qiao, Y. et al. Mechanistic support for combined MET and AR blockade in castration-resistant prostate cancer. Neoplasia 18, 1–9 (2016).
Ahronian, L. G. & Corcoran, R. B. Strategies for monitoring and combating resistance to combination kinase inhibitors for cancer therapy. Genome Med. 9, 37 (2017).
Smith, D. C. et al. Cabozantinib in patients with advanced prostate cancer: results of a phase II randomized discontinuation trial. J. Clin. Oncol. 31, 412–419 (2013).
Smith, M. et al. Phase III study of cabozantinib in previously treated metastatic castration-resistant prostate cancer: COMET-1. J. Clin. Oncol. 34, 3005–3013 (2016).
Hudkins, R. L. et al. Synthesis and biological profile of the pan-vascular endothelial growth factor receptor/tyrosine kinase with immunoglobulin and epidermal growth factor-like homology domains 2 (VEGF-R/TIE-2) inhibitor 11-(2-methylpropyl)-12,13-dihydro-2-methyl-8-(pyrimidin-2-ylamino)-4H-indazolo[5, 4-a]pyrrolo[3,4-c]carbazol-4-one (CEP-11981): a novel oncology therapeutic agent. J. Med. Chem. 55, 903–913 (2012).
Pili, R., Carducci, M., Brown, P. & Hurwitz, H. An open-label study to determine the maximum tolerated dose of the multitargeted tyrosine kinase inhibitor CEP-11981 in patients with advanced cancer. Invest. New Drugs 32, 1258–1268 (2014).
Shisheva, A. PIKfyve: partners, significance, debates and paradoxes. Cell Biol. Int. 32, 591–604 (2008).
Gayle, S. et al. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood 129, 1768–1778 (2017).
Baird, A. M. et al. IL-23R is epigenetically regulated and modulated by chemotherapy in non-small cell lung cancer. Front. Oncol. 3, 162 (2013).
Bonolo De Campos, C. et al. Identification of PIKfyve kinase as a target in multiple myeloma. Haematologica 105, 1641–1649 (2019).
Levy, J. M. M., Towers, C. G. & Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542 (2017).
Noman, M. Z. et al. Inhibition of Vps34 reprograms cold into hot inflamed tumors and improves anti-PD-1/PD-L1 immunotherapy. Sci. Adv. 6, eaax7881 (2020).
Mgrditchian, T. et al. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl Acad. Sci. USA 114, E9271–E9279 (2017).
Wei, H. et al. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev. 25, 1510–1527 (2011).
Yamamoto, K. et al. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581, 100–105 (2020).
Beer, T. M. et al. Randomized, double-blind, phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J. Clin. Oncol. 35, 40–47 (2017).
Kwon, E. D. et al. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 15, 700–712 (2014).
Wheeler, D. L., Iida, M. & Dunn, E. F. The role of Src in solid tumors. Oncologist 14, 667–678 (2009).
Nagasawa, J. et al. Novel HER2 selective tyrosine kinase inhibitor, TAK-165, inhibits bladder, kidney and androgen-independent prostate cancer in vitro and in vivo. Int. J. Urol. 13, 587–592 (2006).
Harshman, L. C. et al. An investigator-initiated phase I study of crizotinib in combination with enzalutamide in metastatic castration-resistant prostate cancer (mCRPC) before or after progression on docetaxel. J. Clin. Oncol. 34, e16509 (2016).
Tripathi, A. et al. Dual blockade of c-MET and the androgen receptor in metastatic castration-resistant prostate cancer: a phase I study of concurrent enzalutamide and crizotinib. Clin. Cancer Res. 26, 6122–6131 (2020).
Hickman, J. A. et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol. J. 9, 1115–1128 (2014).
Harma, V. et al. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS ONE 5, e10431 (2010).
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015).
Yang, Z. J., Chee, C. E., Huang, S. & Sinicrope, F. A. The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 10, 1533–1541 (2011).
Klionsky, D. et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222 2016).
Marx, V. Autophagy: eat thyself, sustain thyself. Nat. Methods 12, 1121–1125 (2015).
Kim, J. & Klionsky, D. J. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69, 303–342 (2000).
Miller, W. T. Tyrosine kinase signaling and the emergence of multicellularity. Biochim. Biophys. Acta 1823, 1053–1057 (2012).
Kaizuka, T. et al. An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 (2016).
Poillet-Perez, L. et al. Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573 (2018).
Kraya, A. A. et al. Identification of secreted proteins that reflect autophagy dynamics within tumor cells. Autophagy 11, 60–74 (2015).
Liu, M., Guo, S. & Stiles, J. K. The emerging role of CXCL10 in cancer. Oncol. Lett. 2, 583–589 (2011).
Harlin, H. et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 69, 3077–3085 (2009).
Bronger, H. et al. CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br. J. Cancer 115, 553–563 (2016).
Tokunaga, R. et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—a target for novel cancer therapy. Cancer Treat. Rev. 63, 40–47 (2018).
Watson, P. A. et al. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer Res. 65, 11565–11571 (2005).
Serganova, I. et al. Enhancement of PSMA-directed CAR adoptive immunotherapy by PD-1/PD-L1 blockade. Mol. Ther. Oncolytics 4, 41–54 (2017).
Rockenfeller, P. et al. Phosphatidylethanolamine positively regulates autophagy and longevity. Cell Death Differ. 22, 499–508 (2015).
Sharma, G. et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy 15, 1694–1718 (2019).
Gayle, S. et al. B-cell non-Hodgkin lymphoma: selective vulnerability to PIKFYVE inhibition. Autophagy 13, 1082–1083 (2017).
Efe, J. A., Botelho, R. J. & Emr, S. D. The Fab1 phosphatidylinositol kinase pathway in the regulation of vacuole morphology. Curr. Opin. Cell Biol. 17, 402–408 (2005).
Ciciola, P., Cascetta, P., Bianco, C., Formisano, L. & Bianco, R. Combining immune checkpoint inhibitors with anti-angiogenic agents. J. Clin. Med. 9, 675 (2020).
Sbrissa, D., Ikonomov, O. C. & Shisheva, A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274, 21589–21597 (1999).
Jefferies, H. B. et al. A selective PIKfyve inhibitor blocks PtdIns(3,5)P2 production and disrupts endomembrane transport and retroviral budding. EMBO Rep. 9, 164–170 (2008).
Choy, C. H. et al. Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. J. Cell Sci. 131, jcs213587 (2018).
Nguyen, H. G. et al. Targeting autophagy overcomes enzalutamide resistance in castration-resistant prostate cancer cells and improves therapeutic response in a xenograft model. Oncogene 33, 4521–4530 (2014).
Yang, S. et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 25, 717–729 (2011).
Saleem, A. et al. Effect of dual inhibition of apoptosis and autophagy in prostate cancer. Prostate 72, 1374–1381 (2012).
Santanam, U. et al. Atg7 cooperates with Pten loss to drive prostate cancer tumor growth. Genes Dev. 30, 399–407 (2016).
Poillet-Perez, L. et al. Autophagy promotes growth of tumors with high mutational burden by inhibiting a T-cell immune response. Nat. Cancer 1, 923–934 (2020).
Antonarakis, E. S. et al. Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 38, 395–405 (2020).
Abida, W. et al. Analysis of the prevalence of microsatellite instability in prostate cancer and response to immune checkpoint blockade. JAMA Oncol. 5, 471–478 (2019).
Antonarakis, E. S. et al. Clinical features and therapeutic outcomes in men with advanced prostate cancer and DNA mismatch repair gene mutations. Eur. Urol. 75, 378–382 (2019).
Antonarakis, E. S. et al. CDK12-altered prostate cancer: clinical features and therapeutic outcomes to standard systemic therapies, poly (ADP-ribose) polymerase inhibitors, and PD-1 inhibitors. JCO Precis. Oncol 4, 370–381 (2020).
Wu, Y. M. et al. Inactivation of CDK12 delineates a distinct immunogenic class of advanced prostate cancer. Cell 173, 1770–1782.e14 (2018).
Chen, C. D. et al. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 10, 33–39 (2004).
Wang, L. et al. VSTM2A overexpression is a sensitive and specific biomarker for mucinous tubular and spindle cell carcinoma (MTSCC) of the kidney. Am. J. Surg. Pathol. 42, 1571–1584 (2018).
Bernard, A. et al. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 25, 546–555 (2015).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Jafari, R. et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 9, 2100–2122 (2014).
Palanisamy, N. et al. The MD Anderson prostate cancer patient-derived xenograft series (MDA PCa PDX) captures the molecular landscape of prostate cancer and facilitates marker-driven therapy development. Clin. Cancer Res. 26, 4933–4946 (2020).
Yu, J. L. et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat. Med. 27, 152–164 (2021).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Afshinnia, F. et al. Lipidomic signature of progression of chronic kidney disease in the chronic renal insufficiency cohort. Kidney Int. Rep. 1, 256–268 (2016).
Bligh, E. G. & Dyer, W. J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 (1959).
Cajka, T. & Fiehn, O. LC-MS-based lipidomics and automated identification of lipids using the LipidBlast in-silico MS/MS library. Methods Mol. Biol. 1609, 149–170 (2017).
Acknowledgements
We thank M. Trierweiler and S. Zelenka-Wang for histological sample processing and IHC, as well as undergraduate student K. Johnson for technical assistance. This work was supported by a Prostate Cancer Foundation Challenge Award, NCI Prostate SPORE Grant P50CA186786, Department of Defense PC130151P1 (to N.M.N. and A.M.C.) and NIH grant GM131919 (to D.J.K.). A.M.C. is an NCI Outstanding Investigator (R35CA231996), Howard Hughes Medical Institute Investigator, A. Alfred Taubman Scholar and American Cancer Society Professor. Y.Q. and J.C.T. are supported by Prostate Cancer Foundation Young Investigator awards. L.X. is supported by a Department of Defense Postdoctoral Award (W81XWH-16-1-0195). E.-L.E. was supported by the Academy of Finland.
Author information
Authors and Affiliations
Contributions
Y.Q. and A.M.C. participated in the planning, initiation and overall analysis of data, as well as writing, reviewing and editing of the manuscript. Y.Q., S.A.S., A.D.D., N.B.H., P.D. and S.M. performed the in vitro and in vivo experiments. J.C.T., J.E.C., K.J. and A.X. participated in the in vivo experiments. Y.Q., T.R. and T.S. participated in the lipidomics experimental design and data analysis. Z.W. and K.D. participated in execution of the chemical synthesis of ESK981. L.W., X.-M.W. and J.S. performed the histological sample preparation, staining and interpretation of RNA ISH results. L.X. helped with the CRISPR Atg5 knockout design. X.W. assisted with the ELISA experiments. X.C., F.S., R.W. and J.N.V. performed the RNA-seq library preparation, sequencing and data analysis. J.Y., I.K. and J.E.C. participated in the flow cytometry analysis. A.B. and D.J.K. participated in the yeast experiments and data interpretation. E.-L.E. performed the electron microscopy analysis. E.-L.E. and D.J.K. participated in the autophagy data interpretation. N.M.N. provided the PDX models. S.J.E. participated in writing and preparation of the manuscript. W.Z. participated in the immune checkpoint blockade experimental design and data interpretation. E.F.-S., E.I.H. and A.M.C. provided project oversight for clinical trial design and review based on the interpretation of the preclinical data.
Corresponding author
Ethics declarations
Competing interests
The University of Michigan has filed a disclosure on the findings based on this study. A.M.C. and Y.Q. are named as co-inventors on the disclosure. Esanik Therapeutics licensed ESK981 from Teva Pharmaceuticals. A.M.C. is a co-founder of Esanik Therapeutics and serves on its scientific advisory board. Neither Esanik Therapeutics nor Teva Pharmaceuticals was involved in the design or approval of this study, nor was this study funded by them. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Cancer thanks Cory Abate-Shen, Thorbald van Hall and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 ESK981 blocks cell growth, induces cell cycle arrest, and decreases cellular invasion.
a-b, Representative crystal violet staining for a long-term survival assay of a panel of prostate cell lines at various concentrations of ESK981, crizotinib, or cabozantinib. c, Cell cycle analysis was measured after 72 hours of increasing concentrations of ESK981 treatment in indicated prostate cancer cell lines. Ctrl, control. d, Cell cycle analysis of VCaP cells that were treated with the indicated compounds for 72 hours. Cabo, cabozantinib; Crizo, crizotinib; Enza, enzalutamide; ESK, ESK981. e, Matrigel invasion assay of various prostate cancer cell lines that were treated with the indicated concentrations of ESK981. The percentage invasion was quantified with a fluorescent plate reader. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated.
Extended Data Fig. 2 ESK981 inhibits the growth of diverse preclinical models of prostate cancer in vivo.
a, Schematic illustration of the VCaP CRPC mouse xenograft experimental design. To generate castration-resistant VCaP, parental VCaP cells were injected subcutaneously into both flanks of intact male mice. When average VCaP tumors reached 200 mm3, mice were surgically castrated and VCaP tumors regressed due to loss of androgen. Castration-resistant VCaP tumors developed as VCaP tumors grew back to the size of pre-castration. Castration-resistant VCaP tumors were then randomized into three groups and treated with vehicle, 30 mg/kg, or 60 mg/kg ESK981 p.o., oral gavage. b, Representative IHC images for proliferation marker Ki67 are shown after treatment with the indicated drugs for five days in VCaP tumors (left). Quantification of positive Ki67 percentage is shown on the right (right). Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM. N = 4 tumors per group. P-value indicated. c, Representative individual tumors from vehicle and ESK981 groups in AR+ and ERG+ prostate PDX MDA-PCa-146-12 (left). Representative IHC showing Ki67 staining for vehicle and 30 mg/kg ESK981 groups of MDA-PCa-146-12 tumors (right) from three independent experiments. d, Representative individual tumors from vehicle and ESK981 groups of DU145 tumors (left). Representative IHC showing Ki67 staining for the vehicle and 30 mg/kg ESK981 groups of DU145 tumors (right) from three independent experiments.
Extended Data Fig. 3 Renal function, liver function, and histopathological evaluation of ESK981-treated xenografts.
a, Castration-resistant VCaP tumors were established according to Extended Data Fig. 2a. Tumor-bearing mice were divided into vehicle and ESK981 50 mg/kg groups, and tumor volumes were monitored twice per week for six weeks. Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM at day 25. N = number of tumors and P-value indicated. b, The percent body weights of VCaP tumor-bearing mice were monitored daily throughout this study. Data were presented as mean ± SEM. N = number of mice. c, The weight of VCaP tumors from vehicle (n = 18 tumors) and ESK981 50 mg/kg (n = 10 tumors) were measured at the end of this study. Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM. P-value indicated. d, Blood chemistry was evaluated for renal and liver functions in non-tumor-bearing and VCaP tumor-bearing mice in vehicle and 50 mg/kg ESK981 treatment groups. e, Representative histological sections showing H&E staining for various organs taken from vehicle- or ESK981-treated mice from three independent experiments. f, Representative histological sections showing H&E staining for tumors taken from vehicle- or ESK981-treated mice from three independent experiments.
Extended Data Fig. 4 ESK981 robustly induces autophagosome levels and is dependent on ATG5 for its effects.
a, DU145 cells with the indicated drug treatment for 24 hours. Autophagosome induction activity was visualized by CYTO-ID® assay from three independent experiments. Rapa, rapamycin. b, VCaP cells were treated with 300 nM ESK981 for the indicated time points, and LC3 protein levels were assessed by western blot from three independent experiments. c, VCaP cells were treated with ESK981 (ESK), crizotinib (Crizo), and cabozantinib (Cabo) at the indicated concentrations. Protein levels of LC3 were examined after 24 hours of treatment from three independent experiments. d, Protein levels of Atg8 in yeast prd5Δ cells after ESK981 (ESK) or cabozantinib (Cabo) treatment under nitrogen deprivation conditions. NT, no treatment. Data were analyzed by two-tailed unpaired t test from four independent experiments and presented as mean ± SEM. P value indicated. e, Protein levels of indicated protein post various siRNA knockdown in VCaP and LNCaP cells with or without 300 nM ESK981 or 1 µM sunitinib treatment for 24 hours from three independent experiments.
Extended Data Fig. 5 ESK981 upregulates CXCL10 expression in human prostate cancer cells and inhibits autophagy in murine Myc-CaP prostate cancer cells.
(a) CXCL10 protein levels measured by ELISA in conditioned media from VCaP cells treated with ESK981 or various autophagy inducers for 24 hours. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated. (b) CXCL10 mRNA levels measured by quantitative PCR (qPCR) in VCaP, PC3, and DU145 cells with the indicated treatment for 24 hours. IFNγ, interferon gamma. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated. (c) IC50 of ESK981, crizotinib, and cabozantinib determined in Myc-CaP cells. (d) Protein levels of LC3 after 50 nM, 100 nM, and 300 nM ESK981 treatment for 24 hours in Myc-CaP cells from three independent experiments. (e) Ratio of GFP/RFP signal in Myc-CaP GFP-LC3-RFP-LC3∆G stable expressing cells with the indicated treatment for 24 hours. Data were analyzed by two-tailed unpaired t test from four independent experiments and presented as mean ± SEM. P-value indicated.
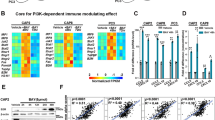
Extended Data Fig. 6 Atg5 deletion blocks ESK981-induced vacuolization and CXCL10-mediated immune response.
(a) Myc-CaP wild-type (WT) and Atg5 knockout (Atg5 KO) cells were treated with increasing concentrations of ESK981 for 24 hours. Atg5 and LC3 levels were assessed by western blot from three independent experiments. GAPDH served as a loading control. (b) Representative morphology of vacuolization in Myc-CaP wild-type (WT) and Atg5 knockout (Atg5 KO) cells after treatment with control or 100 nM ESK981 for 24 hours from three independent experiments. (c) Autophagosome content of Myc-CaP WT and Atg5 KO cells were measured by CYTO-ID® assay after being treated with increasing concentrations of ESK981 for 24 hours. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated. (d) Mouse cytokine array using Myc-CaP WT and Atg5 KO cell supernatant after treatment with 10 ng/ml mouse interferon gamma (mIFNγ) or mIFNγ + 100 nM ESK981 for 24 hours. Differential expression candidate dots are highlighted by boxes. (e) Mouse CXCL10 protein levels were measured by ELISA in Myc-CaP WT and Atg5 KO conditioned medium with the indicated treatment for 24 hours. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated. (f) mRNA levels of Cxcl10 and Cxcl9 were measured by qPCR in Myc-CaP WT and Atg5 KO cells with 50 nM or 100 nM ESK981 and 10 ng/ml mIFNγ treatment for 24 hours. Data were analyzed by two-tailed unpaired t test from three independent experiments and presented as mean ± SEM. P-value indicated.
Extended Data Fig. 7 Transcriptomic analysis of Myc-CaP tumors treated with ESK981 in combination with anti-PD-1 immunotherapy in FVB mice.
(a) Principal Component Analysis (PCA) of individual Myc-CaP tumors from indicated treatment groups based on variance-stabilizing transformation (vst) of read-count data. The vehicle and ESK981+anti-PD-1 combination groups form a relatively distinct cluster based on the first two principal components. (b) Volcano plot of differential gene expression analysis for groups treated with ESK981+anti-PD-1 versus vehicle. The horizontal dashed line corresponds to the FDR = 0.05. The vertical dashed lines correspond to log2FC >= 1 (up-regulation) or log2FC <= -1 (down-regulation). (c) Mouse Gene Set Enrichment Analysis (GSEA) with biological process gene ontology for groups treated with ESK981+anti-PD-1 versus vehicle. Top 10 gene sets are ordered by normalized enrichment score (NES). The top enriched categories are relevant to immune responses and inflammation. (d) Heatmap representation of top differentially expressed genes in groups treated with ESK981+anti-PD-1 versus vehicle (FDR <= 0.01, up or down-regulated by at least 2-fold). (e) Fragments per kilobase of exon model per million reads mapped (FPKM) of indicated targets from individual Myc-CaP tumors treated with vehicle (n = 10 tumors), ESK981 (n = 8 tumors), anti-PD-1 (n = 7 tumors), or ESK981+anti-PD-1 (n = 8 tumors). Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM. P-value indicated.
Extended Data Fig. 8 ESK981 induces autophagosome formation and upregulates Cxcl10 expression in various murine cancer cell lines.
a, IC50 from cell viability assays of ESK981 in murine cancer cells of lung (Ae17, LLC), melanoma (B16F10), ovarian (ID8), pancreas (PAN02), renal (Renca), prostate (TRAMP-C2), and breast (4T1) lineages. Data were plotted as mean ± SEM from three independent experiments. b, Autophagosome content measured by CYTO-ID in indicated cell lines treated with control (Ctrl) or 300 nM ESK981 for 24 hours. Data were analyzed by two-tailed unpaired t test from four independent experiments and presented as mean ± SEM. P-value indicated. c, mRNA level of Cxcl10 in indicated cell lines treated with 10 ng/ml mIFNγ or mIFNγ plus 300 nM ESK981 for 24 hours. Data were analyzed by two-tailed unpaired t test from three (PAN02, Ae17) or four (ID8, B16F10, Renca, 4T1, TRAMP-C2, LLC) independent experiments and presented as mean ± SEM. P-value indicated.
Extended Data Fig. 9 ESK981 sensitizes the murine breast cancer 4T1 model to anti-PD-1 immunotherapy.
a, Bioluminescent signaling images showing dorsal and ventral views of individual 4T1 tumor-bearing mice from indicated treatment groups. b, Bioluminescent quantification of total tumor burden from individual mice treated with vehicle (n = 5 mice), anti-PD-1 (n = 4 mice), ESK981 15 mg/kg (n = 5 mice), ESK981+anti-PD-1 (n = 5 mice). Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM. P-value indicated. c, Overall survival of 4T1-bearing mice treated with either anti-PD-1 (n = 15 mice) or ESK981 and anti-PD-1 combination (n = 15 mice).
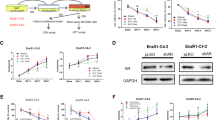
Extended Data Fig. 10 PIKfyve mediates a cellular vacuolization morphology in human prostate cancer cells and murine cancer cells, and Pikfyve loss induces accumulation of autophagosomes in various murine cancer cells.
a, Morphology of DU145 and PC3 cells after siNC, siPIKFYVE, siPIP5K1C, or siPIK3CA transfection from three independent experiments. b, mRNA levels of PIKFYVE, PIP5K1C, and PIK3CA were measured by qPCR after siRNA knockdown of indicated targets in DU145 and PC3 cells. Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM from three independent experiments. P-value indicated. c, Morphological changes of TRAMP-C2, ID8, and Ae17 cells after siNC or siPikfyve transfection from three independent experiments. d, Autophagosome induction activity measured with CYTO-ID® assay in TRAMP-C2, ID8, and Ae17 cells after siRNA knockdown of Pikfyve. Data were analyzed by two-tailed unpaired t test and presented as mean ± SEM from four (TRAMP-C2 and ID8) and six (Ae17) independent experiments. P-value indicated.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 5
Unprocessed western blots.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Fig. 7
Unprocessed western blots.
Source Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 4
Unprocessed western blots.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed western blots.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Qiao, Y., Choi, J.E., Tien, J.C. et al. Autophagy inhibition by targeting PIKfyve potentiates response to immune checkpoint blockade in prostate cancer. Nat Cancer 2, 978–993 (2021). https://doi.org/10.1038/s43018-021-00237-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-021-00237-1
This article is cited by
-
The role of autophagy in prostate cancer and prostatic diseases: a new therapeutic strategy
Prostate Cancer and Prostatic Diseases (2024)
-
The role of HIF in angiogenesis, lymphangiogenesis, and tumor microenvironment in urological cancers
Molecular Biology Reports (2024)
-
Beyond PI3Ks: targeting phosphoinositide kinases in disease
Nature Reviews Drug Discovery (2023)
-
Investigation and verification of GIMAP6 as a robust biomarker for prognosis and tumor immunity in lung adenocarcinoma
Journal of Cancer Research and Clinical Oncology (2023)
-
Targeting autophagy in prostate cancer: preclinical and clinical evidence for therapeutic response
Journal of Experimental & Clinical Cancer Research (2022)