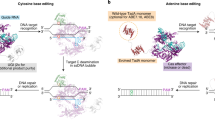
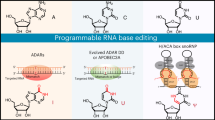
Abstract
Base editing — the introduction of single-nucleotide variants (SNVs) into DNA or RNA in living cells — is one of the most recent advances in the field of genome editing. As around half of known pathogenic genetic variants are due to SNVs, base editing holds great potential for the treatment of numerous genetic diseases, through either temporary RNA or permanent DNA base alterations. Recent advances in the specificity, efficiency, precision and delivery of DNA and RNA base editors are revealing exciting therapeutic opportunities for these technologies. We expect the correction of single point mutations will be a major focus of future precision medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Sun, H. & Yu, G. New insights into the pathogenicity of non-synonymous variants through multi-level analysis. Sci. Rep. 9, 1–11 (2019).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Katsonis, P. et al. Single nucleotide variations: biological impact and theoretical interpretation. Protein Sci. 23, 1650–1666 (2014).
Zhang, F. & Lupski, J. R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 24, R102–R110 (2015).
Kircher, M. et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46, 310–315 (2014).
Bertucci, F. et al. Genomic characterization of metastatic breast cancers. Nature 569, 560–564 (2019).
Smith, C. et al. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol. Ther. 23, 570–577 (2015).
Komor, A. C., Badran, A. H. & Liu, D. R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell 168, 20–36 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). This landmark paper reports the mechanistic elucidation of SpCas9 for programmable double-stranded DNA break introduction, demonstrating its potential for subsequent use as a genome editing agent.
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Martin, R. M. et al. Highly efficient and marker-free genome editing of human pluripotent stem cells by CRISPR–Cas9 RNP and AAV6 donor-mediated homologous recombination. Cell Stem Cell 24, 821–828 (2019).
Paquet, D. et al. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129 (2016).
Richardson, C. D., Ray, G. J., DeWitt, M. A., Curie, G. L. & Corn, J. E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR–Cas9 using asymmetric donor DNA. Nat. Biotechnol. 34, 339–344 (2016).
Yu, C. et al. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell 16, 142–147 (2015).
Lin, S., Staahl, B. T., Alla, R. K. & Doudna, J. A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife 3, e04766 (2014).
Song, J. et al. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 7, 10548 (2016).
Pinder, J., Salsman, J. & Dellaire, G. Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43, 9379–9392 (2015).
Yeh, C. D., Richardson, C. D. & Corn, J. E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 21, 1468–1478 (2019).
Burmistrz, M., Krakowski, K. & Krawczyk-Balska, A. RNA-targeting CRISPR–Cas systems and their applications. Int. J. Mol. Sci. 21, 1122 (2020).
Abudayyeh, O. O. et al. RNA targeting with CRISPR–Cas13. Nature 550, 280–284 (2017).
Gaj, T., Gersbach, C. A. & Barbas III, C. F. ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013).
Gupta, R. M. & Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR–Cas9. J. Clin. Invest. 124, 4154–4161 (2014).
Joung, J. K. & Sander, J. D. TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55 (2013).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). This paper reports the development of the first DNA base editors, BE1, BE2 and BE3, capable of installing C•G to T•A base pair conversions.
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014).
Kunz, C., Saito, Y. & Schär, P. Mismatched repair: variations on a theme. Cell. Mol. Life Sci. 66, 1021–1038 (2009).
Fukui, K. DNA mismatch repair in eukaryotes and bacteria. J. Nucleic Acids 2010, 260512 (2010).
Yang, L. et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 7, 1–12 (2016).
Mok, B. Y. et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583, 631–637 (2020). This paper is the initial report of DdCBE, the first base editor developed for mitochondrial base editing.
Landrum, M. J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44, D862–D868 (2016).
Gaudelli, N. M. et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). This paper reports the development of the first ABEs and the most widely used variant, ABE7.10, capable of installing A•T to G•C base pair conversions.
Shi, K. et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 24, 131–139 (2017).
Grünewald, J. et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. 37, 1041–1048 (2019).
Rallapalli, K. L., Komor, A. C. & Paesani, F. Computer simulations explain mutation-induced effects on the DNA editing by adenine base editors. Sci. Adv. 6, eaaz2309 (2020).
Woolf, T. M., Chase, J. M. & Stinchcomb, D. T. Toward the therapeutic editing of mutated RNA sequences. Biochemistry 92, 8298–8302 (1995). This paper reports the first instance of targeted RNA base editing using complementary RNA oligonucleotides and endogenous ADAR.
Chattopadhyay, S., Garcia-Mena, J., DeVito, J., Wolska, K. & Das, A. Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage λ. Proc. Natl Acad. Sci. USA 92, 4061–4065 (1995).
Fukuda, M. et al. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci. Rep. 7, 8–19 (2017).
Wettengel, J., Reautschnig, P., Geisler, S., Kahle, P. J. & Stafforst, T. Harnessing human ADAR2 for RNA repair — recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res. 45, 2797–2808 (2017).
Vogel, P., Schneider, M. F., Wettengel, J. & Stafforst, T. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew. Chem. Int. Ed. Engl. 53, 6267–6271 (2014).
Schneider, M. F., Wettengel, J., Hoffmann, P. C. & Stafforst, T. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res. 42, e87 (2014).
Stafforst, T. & Schneider, M. F. An RNA–deaminase conjugate selectively repairs point mutations. Angew. Chem. Int. Ed. Engl. 51, 11166–11169 (2012).
Rees, H. A. & Liu, D. R. Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018).
Vogel, P. et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018).
Montiel-Gonźalez, M. F., Vallecillo-Viejo, I. C. & Rosenthal, J. J. C. An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res. 44, e157 (2016).
Montiel-Gonzalez, M. F., Vallecillo-Viejo, I., Yudowski, G. A. & Rosenthal, J. J. C. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl Acad. Sci. USA 110, 18285–18290 (2013).
Vogel, P., Hanswillemenke, A. & Stafforst, T. Switching protein localization by site-directed RNA editing under control of light. ACS Synth. Biol. 6, 1642–1649 (2017).
Hanswillemenke, A., Kuzdere, T., Vogel, P., Jékely, G. & Stafforst, T. Site-directed RNA editing in vivo can be triggered by the light-driven assembly of an artificial riboprotein. J. Am. Chem. Soc. 137, 15875–15881 (2015).
Merkle, T. et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37, 133–138 (2019).
Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019).
Cox, D. B. T. et al. RNA editing with CRISPR–Cas13. Science 358, 1019–1027 (2017). This paper reports the first instance of Cas-derived RNA base editors, resulting in A-to-I base editing.
Kuttan, A. & Bass, B. L. Mechanistic insights into editing-site specificity of ADARs. Proc. Natl Acad. Sci. USA 109, E3295–E3304 (2012).
Wang, Y., Havel, J. & Beal, P. A. A phenotypic screen for functional mutants of human adenosine deaminase acting on RNA 1. ACS Chem. Biol. 10, 2512–2519 (2015).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Nishida, K. et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353, aaf8729 (2016).
Kim, D. et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 35, 475–480 (2017).
Liang, P. et al. Genome-wide profiling of adenine base editor specificity by EndoV-seq. Nat. Commun. 10, 67 (2019).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR–Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Slaymaker, I. M. et al. Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88 (2016).
Yin, J. et al. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. 5, 18 (2019).
Kleinstiver, B. P. et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495 (2016).
Chen, J. S. et al. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 550, 407–410 (2017).
Casini, A. et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 36, 265–271 (2018).
Lee, J. K. et al. Directed evolution of CRISPR–Cas9 to increase its specificity. Nat. Commun. 9, 3048 (2018).
Vakulskas, C. A. et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224 (2018).
Rees, H. A. et al. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 8, 15790 (2017).
Xu, W. et al. Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice. BMC Plant. Biol. 19, 511 (2019).
Park, S. & Beal, P. A. Off-target editing by CRISPR-guided DNA base editors. Biochemistry 58, 3727–3734 (2019).
Zuo, E. et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019).
Jin, S. et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019).
Kim, D., Kim, D.-E., Lee, G., Cho, S.-I. & Kim, J. S. Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 37, 430–435 (2019).
Hua, K., Tao, X., Yuan, F., Wang, D. & Zhu, J. K. Precise A·T to G·C base editing in the rice genome. Mol. Plant. 11, 627–630 (2018).
Yan, F. et al. Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice. Mol. Plant 11, 631–634 (2018). Together with Hua et al. (2018), this paper reported the gRNA-independent off-target DNA activity of CBE, but not ABE.
Kang, B. C. et al. Precision genome engineering through adenine base editing in plants. Nat. Plants 4, 427–431 (2018).
Doman, J. L., Raguram, A., Newby, G. A. & Liu, D. R. Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. 38, 620–628 (2020).
Yu, Y. et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun. 11, 2052 (2020). Together with Doman et al. (2020), this paper reports next-generation CBEs with decreased gRNA-independent off-target DNA editing.
Gaudelli, N. M. et al. Directed evolution of adenine base editors with increased activity and therapeutic application. Nat. Biotechnol. 38, 892–900 (2020).
Richter, M. F. et al. Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol. 38, 883–891 (2020). Together with Gaudelli et al. (2020), this paper reports an eighth-generation ABE with improved on-target editing efficiencies.
Grünewald, J. et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019).
Rees, H. A., Wilson, C., Doman, J. L. & Liu, D. R. Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 5, eaax5717 (2019).
Zhou, C. et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278 (2019).
Mathony, J. et al. Computational design of anti-CRISPR proteins with improved inhibition potency. Nat. Chem. Biol. 16, 725–730 (2020).
Jiang, F. et al. Temperature-responsive competitive inhibition of CRISPR–Cas9. Mol. Cell 73, 601–610 (2019).
Pawluk, A., Davidson, A. R. & Maxwell, K. L. Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol. 16, 12–17 (2018).
Hwang, S. & Maxwell, K. L. Meet the anti-CRISPRs: widespread protein inhibitors of CRISPR–Cas systems. Cris. J. 2, 23–30 (2019).
Maji, B. et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR–Cas9. Cell 177, 1067–1079 (2019).
Richter, F. et al. Switchable Cas9. Curr. Opin. Biotech. 48, 119–126 (2017).
Pan, Y. et al. Near-infrared upconversion-activated CRISPR–Cas9 system: a remote-controlled gene editing platform. Sci. Adv. 5, eaav7199 (2019).
Hess, G. T. et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 13, 1036–1042 (2016).
Kim, K. et al. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 35, 435–437 (2017).
Ma, Y. et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035 (2016).
Komor, A. C. et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017). This paper reports further insights into the mechanism of CBEs in cells and presents novel CBE variants with increased on-target efficiency and greater product purity.
Wang, L. et al. Enhanced base editing by co-expression of free uracil DNA glycosylase inhibitor. Cell Res. 27, 1289–1292 (2017).
d’Adda di Fagagna, F., Weller, G. R., Doherty, A. J. & Jackson, S. P. The Gam protein of bacteriophage Mu is an orthologue of eukaryotic Ku. EMBO Rep. 4, 47–52 (2003).
Liu, Z. et al. Highly efficient RNA-guided base editing in rabbit. Nat. Commun. 9, 2717 (2018).
Jiang, W. et al. BE-PLUS: a new base editing tool with broadened editing window and enhanced fidelity. Cell Res. 28, 855–861 (2018).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas9 nucleases with altered PAM specificities. Nature 523, 481–485 (2015).
Kleinstiver, B. P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR–Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015).
Kim, Y. B. et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9–cytidine deaminase fusions. Nat. Biotechnol. 35, 371–376 (2017).
Huang, T. P. et al. Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol. 37, 626–631 (2019).
Nishimasu, H. et al. Engineered CRISPR–Cas9 nuclease with expanded targeting space. Science 361, 1259–1262 (2018).
Hu, J. H. et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556, 57–63 (2018).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR–Cas9 variants. Science 368, 290–296 (2020).
Hua, K., Tao, X. & Zhu, J. K. Expanding the base editing scope in rice by using Cas9 variants. Plant. Biotechnol. J. 17, 499–504 (2019).
Yang, L. et al. Increasing targeting scope of adenosine base editors in mouse and rat embryos through fusion of TadA deaminase with Cas9 variants. Protein Cell 9, 814–819 (2018).
Nishimasu, H. et al. Crystal structure of Staphylococcus aureus Cas9. Cell 162, 1113–1126 (2015).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2019).
Li, X. et al. Base editing with a Cpf1–cytidine deaminase fusion. Nat. Biotechnol. 36, 324–327 (2018).
Strecker, J. et al. Engineering of CRISPR–Cas12b for human genome editing. Nat. Commun. 10, 212 (2019).
Gehrke, J. M. et al. An APOBEC3A–Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977 (2018).
Liu, Z. et al. Efficient base editing with high precision in rabbits using YFE-BE4max. Cell Death Dis. 11, 36 (2020).
Tan, J., Zhang, F., Karcher, D. & Bock, R. Engineering of high-precision base editors for site-specific single nucleotide replacement. Nat. Commun. 10, 439 (2019).
Grünewald, J. et al. A dual-deaminase CRISPR base editor enables concurrent adenine and cytosine editing. Nat. Biotechnol. 38, 861–864 (2020).
Zhang, X. et al. Dual base editor catalyzes both cytosine and adenine base conversions in human cells. Nat. Biotechnol. 38, 856–860 (2020).
Sakata, R. C. et al. Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 38, 865–869 (2020). Together with Grünewald et al. (2020) and Zhang et al. (2020), this paper reports the development and application of dual base editors in mammalian cells.
Li, C. et al. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 38, 875–882 (2020). This paper is the first report of a dual base editor, for use in plants.
Kurt, I. C. et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0609-x (2020).
Chen, L. et al. Precise and programmable C:G to G:C base editing in genomic DNA. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.07.21.213827v1 (2020).
Zhao, D. et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat. Biotechnol. https://doi.org/10.1038/s41587-020-0592-2 (2020).
Liang, P. et al. Effective gene editing by high-fidelity base editor 2 in mouse zygotes. Protein Cell 8, 601–611 (2017).
Ma, Y. et al. Highly efficient and precise base editing by engineered dCas9-guide tRNA adenosine deaminase in rats. Cell Discov. 4, 39 (2018).
Zhang, Y. et al. Programmable base editing of zebrafish genome using a modified CRISPR–Cas9 system. Nat. Commun. 8, 118 (2017).
Tanaka, S. et al. In vivo targeted single-nucleotide editing in zebrafish. Sci. Rep. 8, 11423 (2018).
Li, G. et al. Highly efficient and precise base editing in discarded human tripronuclear embryos. Protein Cell 8, 776–779 (2017).
Liang, P. et al. Correction of β-thalassemia mutant by base editor in human embryos. Protein Cell 8, 811–822 (2017).
Hecker, J. G. Non-viral, lipid-mediated DNA and mRNA gene therapy of the central nervous system (CNS): chemical-based transfection. Methods Mol. Biol. 1382, 307–324 (2016).
Tong, S., Moyo, B., Lee, C. M., Leong, K. & Bao, G. Engineered materials for in vivo delivery of genome-editing machinery. Nat. Rev. Mater. 4, 726–737 (2019).
Bessis, N., GarciaCozar, F. J. & Boissier, M. C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 11, S10–S17 (2004).
Eoh, J. & Gu, L. Biomaterials as vectors for the delivery of CRISPR–Cas9. Biomater. Sci. 7, 1240–1261 (2019).
Li, L., Hu, S. & Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: challenges and opportunities. Biomaterials 171, 207–218 (2018).
Kormann, M. S. D. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–159 (2011).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 31, 898–907 (2013).
Bornelöv, S., Selmi, T., Flad, S., Dietmann, S. & Frye, M. Codon usage optimization in pluripotent embryonic stem cells. Genome Biol. 20, 119 (2019).
Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014).
Koblan, L. W. et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–848 (2018).
Hanson, G. & Coller, J. Translation and protein quality control: codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 19, 20–30 (2018).
Zafra, M. P. et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–896 (2018).
Sebestyén, M. G. et al. Mechanism of plasmid delivery by hydrodynamic tail vein injection. I. Hepatocyte uptake of various molecules. J. Gene Med. 8, 852–873 (2006).
Song, C. Q. et al. Adenine base editing in an adult mouse model of tyrosinaemia. Nat. Biomed. Eng. 4, 125–130 (2020).
Zuris, J. A. et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 33, 73–80 (2015).
Sokołowska, E. & Błachnio-Zabielska, A. U. A critical review of electroporation as a plasmid delivery system in mouse skeletal muscle. Int. J. Mol. Sci. 20, 2776 (2019).
Kaneko, T. & Nakagawa, Y. Genome editing of rodents by electroporation of CRISPR/Cas9 into frozen-warmed pronuclear-stage embryos. Cryobiology 92, 231– 234 (2020).
Webber, B. R. et al. Highly efficient multiplex human T cell engineering without double-strand breaks using Cas9 base editors. Nat. Commun. 10, 5222 (2019).
Park, D. S. et al. Targeted base editing via RNA-guided cytidine deaminases in Xenopus laevis embryos. Mol. Cell 40, 823–827 (2017).
Sasaguri, H. et al. Introduction of pathogenic mutations into the mouse Psen1 gene by base editor and target-AID. Nat. Commun. 9, 2892 (2018).
Ryu, S. M. et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol. 36, 536–539 (2018).
Lin, X. et al. Base editing-mediated splicing correction therapy for spinal muscular atrophy. Cell Res. 548–550 (2020).
Swiech, L. et al. In vivo interrogation of gene function in the mammalian brain using CRISPR–Cas9. Nat. Biotechnol. 33, 102–106 (2015).
Maddalena, A. et al. Triple vectors expand AAV transfer capacity in the retina. Mol. Ther. 26, 524–541 (2018).
Chadwick, A. C., Wang, X. & Musunuru, K. In vivo base editing of PCSK9 (proprotein convertase subtilisin/kexin type 9) as a therapeutic alternative to genome editing. Arterioscler. Thromb. Vasc. Biol. 37, 1741–1747 (2017).
Hakim, C. H. et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight 3, e124297 (2018).
Bak, R. O. & Porteus, M. H. CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep. 20, 750–756 (2017).
Yin, H. et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328–333 (2016).
Zhu, H., Zhang, L. & Tong, S. Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets. Nat. Biomed. Eng. 3, 126–136 (2019).
Katrekar, D., Moreno, A. M., Chen, G., Worlikar, A. & Mali, P. Oligonucleotide conjugated multi-functional adeno-associated viruses. Sci. Rep. 8, 3589 (2018).
Chew, W. L. et al. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat. Methods 13, 868–874 (2016).
Li, C. & Lieber, A. Adenovirus vectors in hematopoietic stem cell genome editing. FEBS Lett. 593, 3623–3648 (2019).
Huang, S. & Kamihira, M. Development of hybrid viral vectors for gene therapy. Biotechnol. Adv. 31, 208–223 (2013).
Maggio, I. et al. Integrating gene delivery and gene-editing technologies by adenoviral vector transfer of optimized CRISPR–Cas9 components. Gene Ther. 27, 209–225 (2020).
Yang, Y. et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 34, 334–338 (2016).
Park, A. et al. Sendai virus, an RNA virus with no risk of genomic integration, delivers CRISPR/Cas9 for efficient gene editing. Mol. Ther. Methods Clin. Dev. 3, 16057 (2016).
Rossidis, A. C. et al. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat. Med. 24, 1513–1518 (2018).
Muruve, D. A. The innate immune response to adenovirus vectors. Hum. Gene Ther. 15, 1157–1166 (2004).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Fry, L. E., Peddle, C. F., Barnard, A. R., McClements, M. E. & Maclaren, R. E. RNA editing as a therapeutic approach for retinal gene therapy requiring long coding sequences. Int. J. Mol. Sci. 21, 777 (2020).
Katrekar, D. et al. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 16, 239–242 (2019).
Wu, Z., Yang, H. & Colosi, P. Effect of genome size on AAV vector packaging. Mol. Ther. 18, 80–86 (2010).
Truong, D. J. et al. Development of an intein-mediated split–Cas9 system for gene therapy. Nucleic Acids Res. 43, 6450–6458 (2015).
Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97–110 (2020). This paper reports split-intein base editors capable of being delivered in vivo with AAVs.
Winter, J. et al. Targeted exon skipping with AAV-mediated split adenine base editors. Cell Discov. 5, 41 (2019).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Lim, C. K. W. et al. Treatment of a mouse model of ALS by in vivo base editing. Mol. Ther. 28, P1177–P1189 (2020).
Yeh, W.-H. et al. In vivo base editing restores sensory transduction and transiently improves auditory function in a mouse model of recessive deafness. Sci. Transl. Med. 12, eaay9101 (2020).
Sack, B. K. & Herzog, R. W. Evading the immune response upon in vivo gene therapy with viral vectors. Curr. Opin. Mol. Ther. 11, 493–503 (2009).
Mingozzi, F. & High, K. A. Immune responses to AAV vectors: overcoming barriers to successful gene therapy. Blood 122, 23–36 (2013).
Charlesworth, C. T. et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 25, 249–254 (2019).
Simhadri, V. L. et al. Prevalence of pre-existing antibodies to CRISPR-associated nuclease Cas9 in the USA population. Mol. Ther. Methods Clin. Dev. 10, 105–112 (2018).
Verdera, H. C., Kuranda, K. & Mingozzi, F. AAV vector immunogenicity in humans: a long journey to successful gene transfer. Mol. Ther. 28, 723–746 (2020).
Boutin, S. et al. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 712, 704–712 (2010).
Wagner, D. L. et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 25, 242–248 (2019).
Cho, S.-W., Lee, J., Carroll, D., Kim, J.-S. & Lee, J. Heritable gene knockout in Caenorhabditis elegans by direct injection of Cas9–sgRNA ribonucleoproteins. Genetics 195, 1177–1180 (2013).
Kim, S., Kim, D., Cho, S. W., Kim, J. & Kim, J. S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019 (2014).
DeWitt, M. A., Corn, J. E. & Carroll, D. Genome editing via delivery of Cas9 ribonucleoprotein. Methods 121–122, 9–15 (2017).
Paix, A., Folkmann, A., Rasoloson, D. & Seydoux, G. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR–Cas9 ribonucleoprotein complexes. Genetics 201, 47–54 (2015).
Liang, X. et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 208, 44–53 (2015).
Yeh, W. H., Chiang, H., Rees, H. A., Edge, A. S. B. & Liu, D. R. In vivo base editing of post-mitotic sensory cells. Nat. Commun. 9, 2184 (2018).
Zeng, J. et al. Therapeutic base editing of human hematopoietic stem cells. Nat. Med. 26, 535–541 (2020).
Gao, X. et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217–221 (2018).
Glass, Z., Lee, M., Li, Y. & Xu, Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 36, 173–185 (2018).
Lee, K. et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 1, 889–901 (2017).
Givens, B. E., Naguib, Y. W., Geary, S. M., Devor, E. J. & Salem, A. K. Nanoparticle based delivery of CRISPR/Cas9 genome editing therapeutics. AAPS J. 20, 108 (2019).
Yin, H. et al. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014).
Chen, Z. et al. Targeted delivery of CRISPR/Cas9-mediated cancer gene therapy via liposome-templated hydrogel nanoparticles. Adv. Funct. Mater. 27, 1703036 (2017).
Getts, D. R., Shea, L. D., Miller, S. D. & King, N. J. C. Harnessing nanoparticles for immune modulation. Trends Immunol. 36, 419–427 (2015).
Walkey, C. D. et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 8, 2439–2455 (2014).
Walkey, C. D., Olsen, J. B., Guo, H., Emili, A. & Chan, W. C. W. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 134, 2139–2147 (2012).
Lima, T., Bernfur, K., Vilanova, M. & Cedervall, T. Understanding the lipid and protein corona formation on different sized polymeric nanoparticles. Sci. Rep. 10, 1129 (2020).
Mirakabad, F. S. T. et al. PLGA-based nanoparticles as cancer drug delivery systems. Asian Pac. J. Cancer Prev. 15, 517–535 (2014).
Chen, K. et al. Cationic polymeric nanoformulation: recent advances in material design for CRISPR/Cas9 gene therapy. Prog. Nat. Sci. Mater. Int. 29, 617–627 (2019).
Xu, C. et al. Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases. Nat. Commun. 9, 4092 (2018).
Liu, Y. et al. Systemic delivery of CRISPR/Cas9 with PEG–PLGA nanoparticles for chronic myeloid leukemia targeted therapy. Biomater. Sci. 6, 1592–1603 (2018).
Lee, B. et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of Fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2, 497–507 (2018).
Langan, R. A. et al. De novo design of bioactive protein switches. Nature 572, 205–210 (2019).
Brunette, T. et al. Modular repeat protein sculpting using rigid helical junctions. Proc. Natl Acad. Sci. USA 117, 8870–8875 (2020).
Jun, H., Wang, X., Bricker, W. P. & Bathe, M. Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 10, 5419 (2019).
Scott, D. A. & Zhang, F. Implications of human genetic variation on CRISPR-based therapeutic genome editing. Nat. Med. 23, 1095–1101 (2017).
June, C. H., O’connor, R. S., Kawalekar, O. U., Ghassemi, S. & Milone, M. C. CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018).
June, C. H. & Sadelain, M. Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018).
Depil, S., Duchateau, P., Grupp, S. A., Mufti, G. & Poirot, L. ‘Off-the-shelf’ allogeneic CAR T cells: development and challenges. Nat. Rev. Drug Discov. 19, 185–199 (2020).
Yang, Y., Jacoby, E. & Fry, T. J. Challenges and opportunities of allogeneic donor-derived CAR T cells. Curr. Opin. Hematol. 22, 509–515 (2015).
Ren, J. et al. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin. Cancer Res. 23, 2255–2266 (2017).
Liu, X. et al. CRISPR–Cas9-mediated multiplex gene editing in CAR-T cells. Cell Res. 27, 154–157 (2017).
Kuscu, C. et al. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat. Methods 14, 710–712 (2017).
Fujioka, M., Okano, H. & Edge, A. S. B. Manipulating cell fate in the cochlea: a feasible therapy for hearing loss. Trends Neurosci. 38, 139–144 (2015).
McLean, W. J. et al. Clonal expansion of Lgr5-positive cells from mammalian cochlea and high-purity generation of sensory hair cells. Cell Rep. 18, 1917–1929 (2017).
Shi, F., Hu, L. & Edge, A. S. B. Generation of hair cells in neonatal mice by β-catenin overexpression in Lgr5-positive cochlear progenitors. Proc. Natl Acad. Sci. USA 110, 13851–13856 (2013).
Verheyen, E. M. & Gottardi, C. J. Regulation of Wnt/β-catenin signaling by protein kinases. Dev. Dynam. 239, 34–44 (2010).
Polakis, P. Wnt signaling and cancer. Genes Dev. 14, 1837–1851 (2000).
Li, L. et al. Advances in nano-based inner ear delivery systems for the treatment of sensorineural hearing loss. Adv. Drug Deliv. Rev. 108, 2–12 (2017).
Mittal, R. et al. Nanoparticle-based drug delivery in the inner ear: current challenges, limitations and opportunities. Artif. Cell Nanomed. Biotechnol. 47, 1312–1320 (2019).
Jawa, V. et al. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. J. Clin. Immunol. 149, 534–555 (2013).
Nowak, K. J. & Davies, K. E. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 5, 872–876 (2004).
Bladen, C. L. et al. The TREAT-NMD DMD Global database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 36, 395–402 (2015).
Echigoya, Y., Lim, K. R. Q., Nakamura, A. & Yokota, T. Multiple exon skipping in the Duchenne muscular dystrophy hot spots: prospects and challenges. J. Pers. Med. 8, 41 (2018).
Crudele, J. M. & Chamberlain, J. S. AAV-based gene therapies for the muscular dystrophies. Hum. Mol. Genet. 28, R102–R107 (2019).
Min, Y.-L., Bassel-Duby, R. & Olson, E. N. CRISPR correction of Duchenne muscular dystrophy. Annu. Rev. Med. 70, 239–255 (2019).
Long, C. et al. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science 345, 1184–1188 (2014).
Trevisan, M., Masi, G. & Palù, G. Genome editing technologies to treat rare liver diseases. Transl. Gastroenterol. Hepatol. 5, 23–23 (2020).
Ebrahimi, A. & Rahim, F. Crigler–Najjar syndrome: current perspectives and the application of clinical genetics. Endoxr. Metab. Immune Disord. Drug Targets 18, 201–211 (2017).
Famulari, E. S. et al. Human liver stem cells express UGT1A1 and improve phenotype of immunocompromised Crigler Najjar syndrome type I mice. Sci. Rep. 10, 887 (2020).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29, 308–311 (2001).
Anzalone, A. V. et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157 (2019). This paper reports the initial development of ‘prime editing’.
Anzalone, A. V., Koblan, L. W. & Liu, D. R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020).
Choi, J. G. et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 23, 627–633 (2016).
Montagna, C. et al. VSV-G-enveloped vesicles for traceless delivery of CRISPR–Cas9. Mol. Ther. Nucleic Acids 12, 453–462 (2018).
Cota-Coronado, A., Díaz-Martínez, N. F., Padilla-Camberos, E. & Díaz-Martínez, N. E. Editing the central nervous system through CRISPR/Cas9 systems. Front. Mol. Neurosci. 12, 110 (2019).
Pardridge, W. M. Molecular Trojan horses for blood–brain barrier drug delivery. Curr. Opin. Pharmacol. 6, 494–500 (2006).
Oakes, B. L. et al. CRISPR–Cas9 circular permutants as programmable scaffolds for genome modification. Cell 176, 254–267 (2019).
Chen, J. S. et al. CRISPR–Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Kleinstiver, B. P. et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Sun, W. et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angew. Chem. Int. Ed. Engl. 54, 12029–12033 (2015).
Guan, X., Luo, Z. & Sun, W. A peptide delivery system sneaks CRISPR into cells. J. Biol. Chem. 293, 17306–17307 (2018).
Hwang, G. H. et al. Web-based design and analysis tools for CRISPR base editing. BMC Bioinforma. 19, 542 (2018).
Dandage, R., Després, P. C., Yachie, N. & Landry, C. R. beditor: a computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing. Genetics 212, 377–385 (2019).
Arbab, M. et al. Determinants of base editing outcomes from target library analysis and machine learning. Cell 182, 1–18 (2020).
Bhagwat, A. M. et al. multicrispr: gRNA design for prime editing and parallel targeting of thousands of targets. Life Sci. Alliance 3, e202000757 (2020).
Hsu, J. Y. et al. PrimeDesign software for rapid and simplified design of prime editing guide RNAs. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.05.04.077750v1 (2020).
Hess, G. T., Tycko, J., Yao, D. & Bassik, M. C. Methods and applications of CRISPR-mediated base editing in eukaryotic genomes. Mol. Cell 68, 26–43 (2017).
Jun, S., Lim, H., Chun, H., Lee, J. H. & Bang, D. Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commun. Biol. 3, 154 (2020).
Kweon, J. et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene 39, 30–35 (2020).
Després, P. C., Dubé, A. K., Seki, M., Yachie, N. & Landry, C. R. Perturbing proteomes at single residue resolution using base editing. Nat. Commun. 11, 1871 (2020).
Hanna, R. E. et al. Massively parallel assessment of human variants with base editor screens. Preprint at bioRxiv https://www.biorxiv.org/content/10.1101/2020.05.17.100818v1 (2020).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Bikard, D. et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR–Cas system. Nucleic Acids Res. 41, 7429–7437 (2013).
Lunshof, J. E. Human germ line editing — roles and responsibilities. Protein Cell 7, 7–10 (2016).
Acknowledgements
E.M.P. is supported by the Molecular Biophysics Training Grant, National Institutes of Health (NIH) Grant T32 GM008326. A.C.K. is partially funded by NIH grant R21 GM135736. G.W.Y. is partially funded by NIH grants EY029166 and NS103172. The authors gratefully acknowledge M. Singh, G. Ciaramella and N. Gaudelli for helpful discussions.
Author information
Authors and Affiliations
Contributions
E.M.P. conceptualized, wrote and edited the manuscript and designed the figures. A.C.K. conceptualized, wrote and edited the manuscript. I.M.S. and G.W.Y. contributed substantially to the content and further reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
A.C.K. is a member of the scientific advisory board (SAB) of Pairwise Plants, and is an equity holder for Pairwise Plants and Beam Therapeutics. I.M.S. is an employee and shareholder of Beam Therapeutics. G.W.Y. is co-founder, member of the Board of Directors, on the SAB, equity holder and paid consultant for Locana and Eclipse BioInnovations. G.W.Y. is a visiting professor at the National University of Singapore. A.C.K.’s and G.W.Y.’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The authors declare no other competing financial interests.
Additional information
Peer review information
Nature Reviews Drug Discovery thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Precision medicine
-
The development of disease prevention and treatment strategies based on a patient’s individual characteristics (that is, genomic sequence).
- Single-nucleotide variants
-
(SNVs). Major cause of genetic diseases, targetable with base editors.
- Guide RNA
-
(gRNA). A short sequence of RNA that recognizes the target DNA region of interest and directs the Cas enzyme to bind for editing to occur (also known as the spacer and single guide RNA).
- Cas9
-
(CRISPR-associated protein 9). The enzyme responsible for DNA double-stranded cutting (wtCas9), single-stranded cutting or nicking (Cas9n), or no DNA cutting (wherein Cas9 is catalytically inactive, dCas9) activity. All three Cas9 variants maintain DNA binding ability.
- Base editors
-
Genome editing tools that allow for the direct, irreversible conversion of target cytosine or adenine bases at a specific genomic locus without relying on double-stranded DNA breaks.
- Protospacer
-
A DNA locus of interest targeted with genome editing agent; base pairs with the guide RNA.
- IUPAC nucleotide codes
-
N = adenine/cytosine/guanine/thymine, R = adenine/guanine, Y = cytosine/thymine, V = adenine/cytosine/guanine (in order of appearance).
- Protospacer adjacent motif
-
(PAM). A variable region on the 5′ or 3′ end of the protospacer, required for Cas protein binding to the target locus. PAM requirements vary among different Cas enzymes (the most widely used Streptococcus pyogenes Cas9 requires an NGG PAM).
- R-loop
-
A tripartite structure consisting of unpaired DNA and a paired DNA:RNA hybrid. Following R-loop formation, the unpaired or single-stranded DNA is accessible for base editing.
- Antisense oligonucleotides
-
(ASOs). Small pieces of DNA or RNA that bind to specific molecules of RNA.
- ADARDD
-
(Deaminase domain of adenosine deaminase acting on RNA enzyme). The first reported case explored for A-to-I RNA base editing.
- Activity window
-
A defined region of single-stranded DNA accessible for base editing activity. Activity windows vary among different base editor variations.
- Ribonucleoprotein complexes
-
(RNPs). Macromolecular structures containing both Cas9 protein and guide RNA molecules.
- IUPAC amino acid codes
-
V = valine, W = tryptophan, R = arginine, A = alanine, K = lysine, G = glycine, Y = tyrosine, E = glutamic acid, P = proline (in order of appearance).
- Prime editing
-
A recently developed genome editing technology that, like base editing, does not rely on double-stranded breaks.
Rights and permissions
About this article
Cite this article
Porto, E.M., Komor, A.C., Slaymaker, I.M. et al. Base editing: advances and therapeutic opportunities. Nat Rev Drug Discov 19, 839–859 (2020). https://doi.org/10.1038/s41573-020-0084-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-020-0084-6
This article is cited by
-
Engineering approaches for RNA-based and cell-based osteoarthritis therapies
Nature Reviews Rheumatology (2024)
-
DNA and RNA base editors can correct the majority of pathogenic single nucleotide variants
npj Genomic Medicine (2024)
-
Crispr-Based Editing of Human Pluripotent Stem Cells for Disease Modeling
Stem Cell Reviews and Reports (2024)
-
C-to-G editing generates double-strand breaks causing deletion, transversion and translocation
Nature Cell Biology (2024)
-
Current trends, limitations and future research in the fungi?
Fungal Diversity (2024)