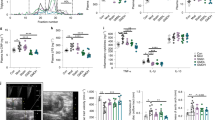
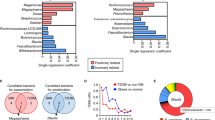
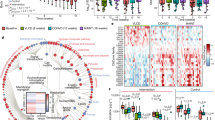
Abstract
The gut microbiome is a malleable microbial community that can remodel in response to various factors, including diet, and contribute to the development of several chronic diseases, including atherosclerosis. We devised an in vitro screening protocol of the mouse gut microbiome to discover molecules that can selectively modify bacterial growth. This approach was used to identify cyclic d,l-α-peptides that remodeled the Western diet (WD) gut microbiome toward the low-fat-diet microbiome state. Daily oral administration of the peptides in WD-fed LDLr−/− mice reduced plasma total cholesterol levels and atherosclerotic plaques. Depletion of the microbiome with antibiotics abrogated these effects. Peptide treatment reprogrammed the microbiome transcriptome, suppressed the production of pro-inflammatory cytokines (including interleukin-6, tumor necrosis factor-α and interleukin-1β), rebalanced levels of short-chain fatty acids and bile acids, improved gut barrier integrity and increased intestinal T regulatory cells. Directed chemical manipulation provides an additional tool for deciphering the chemical biology of the gut microbiome and might advance microbiome-targeted therapeutics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Source data for quantifications either mentioned in the text or shown in graphs are available upon reasonable request from the corresponding authors. RNA-seq data have been deposited in the Gene Expression Omibus under accession GSE104915, and 16S rRNA sequencing reads have been deposited in MG-RAST under accession 93528, 93529 and 93586.
References
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Brown, J. M. & Hazen, S. L. Microbial modulation of cardiovascular disease. Nat. Rev. Microbiol. 16, 171–181 (2018).
Zhao, L. The gut microbiota and obesity: from correlation to causality. Nat. Rev. Microbiol. 11, 639–647 (2013).
Maruvada, P., Leone, V., Kaplan, L. M. & Chang, E. B. The human microbiome and obesity: moving beyond associations. Cell Host Microbe 22, 589–599 (2017).
Khan, M. T., Nieuwdorp, M. & Baeckhed, F. Microbial modulation of insulin sensitivity. Cell Metab. 20, 753–760 (2014).
Pedersen, H. K. et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016).
Sharon, G., Sampson, T. R., Geschwind, D. H. & Mazmanian, S. K. The central nervous system and the gut microbiome. Cell 167, 915–932 (2016).
Turnbaugh, P. J. et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14–16ra14 (2009).
Kau, A. L., Ahern, P. P., Griffin, N. W., Goodman, A. L. & Gordon, J. I. Human nutrition, the gut microbiome and the immune system. Nature 474, 327–336 (2011).
Carmody, R. N. et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17, 72–84 (2015).
Furusawa, Y. et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013).
Rios-Covian, D. et al. Intestinal short chain fatty acids and their link with diet and human health. Front. Microbiol. 7, 185 (2016).
Perry, R. J. et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 534, 213–217 (2016).
Hsiao, E. Y. et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 155, 1451–1463 (2013).
Dodd, D. et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652 (2017).
de Aguiar Vallim, T. Q., Tarling, E. J. & Edwards, P. A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 17, 657–669 (2013).
Wahlstroem, A., Sayin, S. I., Marschall, H.-U. & Baeckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Jia, W., Xie, G. & Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
Schmidt, T. S. B., Raes, J. & Bork, P. The human gut microbiome: from association to modulation. Cell 172, 1198–1215 (2018).
Wang, Z. et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015).
Zhu, W. et al. Precision editing of the gut microbiota ameliorates colitis. Nature 553, 208–211 (2018).
Li, F. et al. Microbiome remodelling leads to inhibition of intestinal Farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4, 2384 (2013).
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R. & Rastall, R. A. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616 (2019).
Shepherd, E. S., DeLoache, W. C., Pruss, K. M., Whitaker, W. R. & Sonnenburg, J. L. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438 (2018).
Chen, M.-L. et al. Resveratrol attenuates trimethylamine-N-oxide (TMAO)-induced atherosclerosis by regulating TMAO synthesis and bile acid metabolism via remodeling of the gut microbiota. MBio 7, e02210–e02215 (2016).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555, 623–628 (2018).
De Santis, P., Morosetti, S. & Rizzo, R. Conformational analysis of regular enantiomeric sequences. Macromolecules 7, 52–58 (1974).
Pavone, V. et al. Regularly alternating l,d-peptides. III. Hexacyclic peptides from valine or phenylalanine. Biopolymers 28, 215–223 (1989).
Ghadiri, M. R., Granja, J. R., Milligan, R. A., McRee, D. E. & Khazanovich, N. Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature 366, 324–327 (1993).
Fernandez-Lopez, S. et al. Antibacterial agents based on the cyclic d,l-ɑ-peptide architecture. Nature 412, 452–456 (2001).
Dartois, V. et al. Systemic antibacterial activity of novel synthetic cyclic peptides. Antimicrob. Agents Chemother. 49, 3302–3310 (2005).
Fletcher, J. T., Finlay, J. A., Callow, M. E., Callow, J. A. & Ghadiri, M. R. A combinatorial approach to the discovery of biocidal six-residue cyclic d,l-ɑ-peptides against the bacteria methicillin-resistant Staphylococcus aureus (MRSA) and E. coli and the biofouling algae Ulva linza and Navicula perminuta. Chem. Eur. J. 13, 4008–4013 (2007).
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006).
Martinez, K. B., Leone, V. & Chang, E. B. Western diets, gut dysbiosis, and metabolic diseases: are they linked? Gut Microbes 8, 130–142 (2017).
Sonnenburg, E. D. et al. Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Baeckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Clark, T. D. et al. Cylindrical beta-sheet peptide assemblies. J. Am. Chem. Soc. 120, 8949–8962 (1998).
Kennedy, E. A., Baldridge, M. T. & King, K. Y. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9, 1534 (2018).
Lundberg, R. et al. Antibiotic-treated versus germ-free rodents for microbiota transplantation studies. Gut Microbes 7, 68–74 (2016).
Zarrinpar, A. et al. Antibiotic-induced microbiome depletion alters metabolic homeostasis by affecting gut signaling and colonic metabolism. Nat. Commun. 9, 2872 (2018).
Miyake, J. H. et al. Transgenic expression of cholesterol-7-alpha-hydroxylase prevents atherosclerosis in C57BL/6J mice. Arterioscler. Thromb. Vasc. Biol. 22, 121–126 (2002).
Li, T. & Chiang, J. Y. L. Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66, 948–983 (2014).
Zhang, Y. et al. FXR deficiency causes reduced atherosclerosis in LDLr−/− mice. Arterioscler. Thromb. Vasc. Biol. 26, 2316–2321 (2006).
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Ross, R. Atherosclerosis–an inflammatory disease. N. Engl. J. Med. 340, 115–126 (1999).
Mazmanian, S. K., Round, J. L. & Kasper, D. L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625 (2008).
Caesar, R., Fak, F. & Backhed, F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J. Intern. Med 268, 320–328 (2010).
Piccirillo, C. A., d’Hennezel, E., Sgouroudis, E. & Yurchenko, E. CD4+FOXP3+ regulatory T cells in the control of autoimmunity: in vivo veritas. Curr. Opin. Immunol. 20, 655–662 (2008).
Ait-Oufella, H. et al. Natural regulatory T cells control the development of atherosclerosis in mice. Nat. Med. 12, 178–180 (2006).
Schiering, C. et al. The alarmin IL-33 promotes regulatory T-cell function in the intestine. Nature 513, 564–568 (2014).
Round, J. L. & Mazmanian, S. K. Inducible FOXP3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl Acad. Sci. USA 107, 12204–12209 (2010).
Sefik, E. et al. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Soroosh, P. et al. Oxysterols are agonist ligands of RORγt and drive Th17 cell differentiation. Proc. Natl Acad. Sci. USA 111, 12163–12168 (2014).
Freigang, S. et al. Fatty acid-induced mitochondrial uncoupling elicits inflammasome-independent IL-1α and sterile vascular inflammation in atherosclerosis. Nat. Immunol. 14, 1045–1053 (2013).
Schloss, P. D. et al. Introducing MOTHUR: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 15, 550 (2014).
Staley, C. et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5, 87 (2017).
Matranga Christian, B. et al. Enhanced methods for unbiased deep sequencing of Lassa and Ebola RNA viruses from clinical and biological samples. Genome Biol. 15, 519 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
O’Leary, N. A. et al. Reference sequence (REFSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745 (2016).
Huson, D. H. et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016).
Hainer Sarah, J. et al. Suppression of pervasive noncoding transcription in embryonic stem cells by ESBAF. Genes Dev. 29, 362–378 (2015).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 12, 323 (2011).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Acknowledgements
We gratefully acknowledge funding from the National Institutes of Health (NHLBI grants R01HL118114 and R01GM125984 to M.R.G., UL1TR001114 supporting B.M. and A.T. and U54GM114833 supporting A.T.), the Skaggs Institute of Chemical Biology and the American Heart Association (postdoctoral fellowships to Y.Z. and P.M.). We thank K. G. Andersen for discussions and protocols and G. Oliveira for technical assistance with sequencing. We thank C. S. Ryan, D. J. Search, R. A. Garcia and D. A. Gordon at Bristol Myers Squibb for carrying out pharmacokinetics and fecal dual isotope cholesterol absorption studies.
Author information
Authors and Affiliations
Contributions
All authors contributed to the design and planning of experiments. P.B.C, B.M., A.L.S. and A.T. planned and carried out bioinformatics analyses of 16S sequencing and RNA-seq data. A.S.B., P.M., G.A.M. and P.S. planned and carried out Treg studies. J.W. and W.C. carried out metabolomics experiments for SCFAs and amino acids. A.F.M.P. and A.S. planned and carried out metabolomics experiments for bile acids. P.B.C., A.S.B., P.M., Y.Z., A.L.S., N.M. and L.J.L. carried out all the other studies described. P.B.C., L.J.L. and M.R.G. wrote the manuscript. All authors provided critical feedback and helped shape the experimental analysis and manuscript. M.R.G. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Changes in gut microbiota composition due to WD feeding in vivo and peptide treatment in vitro.
a, The composition of gut microbiota from fecal samples of LDLr−/− mice after 2-wk feeding of CHD or WD. b, The alpha diversity (biodiversity) of gut microbiota from cecum of LDLr−/− mice cultured in vitro for 20 h with the indicated cyclic peptides (16 μM). Two broad-spectrum antibiotics, 4 μg/mL ampicillin (Amp) and 8 μg/mL chloramphenicol (Chm), were used as positive controls in this assay. The dashed line indicates the biodiversity in the cultured, untreated control sample. c, The ratio of bacteroidetes to firmicutes in the in vitro cultured microbiota samples after cyclic peptide treatment. The dashed line indicates the ratio of bacteroidetes to firmicutes in the cultured, untreated control sample. d, Schematic diagram of the distance-based algorithm for scoring peptides following the in vitro en masse assay. The algorithm considers only the bacteria that increased in abundance under WD feeding compared to CHD feeding (species o, p, etc.). Peptide treatment could cause abundance of those bacteria to further increase (away from the CHD state), to decrease (toward the CHD state), or remain unchanged. Peptides were scored by summing the observed changes in abundance for each of the identified bacteria (Do1, Dp1, etc.), with shifts toward the CHD state being considered positive and shifts away being negative.
Extended Data Fig. 2 Composition of the cultured gut bacteria community from the en masse in vitro screen compared to the uncultured community.
The major observed taxa at the phylum and genus levels are labeled.
Extended Data Fig. 3 Cyclic D,L-α-peptides can affect the growth of individual bacteria or the composition of bacterial communities in vitro.
a, Heatmap showing minimum inhibitory concentration (MIC) values for the screened peptides against selected bacteria in vitro. The MIC for a given peptide against a given bacteria was defined as the lowest concentration of peptide that inhibited >90% of growth; lower MIC values correspond to greater antibacterial activity. The peptide groups shown at the top of the heatmap correspond to those identified from a pairwise comparison of the activities of different cyclic peptides in the en masse assay, each of which affected the microbiota differently from the other groups. Similar to the result from the en masse screen, peptides from different clusters showed the distinct effects against individual gut bacterial species in the MIC assay. Peptide c[wLwKhShK] (1), from peptide group I, broadly affected most of the bacterial species tested to some degree. In contrast, c[wLwReQeR] (11), from peptide group III, differentially affected bacterial species from Firmicutes and Bacteroidetes. Peptide 30 is an N-methylated analog of peptide 11, whereas 31 is a diastereomer of peptide 11. Because these disatereomeric and backbone N-methylated analogs cannot self-assemble into nanotubes, these peptides serve as mode-of-action controls that support the expected mechanism of bacterial growth modulation being dependent on peptide self-assembly and bacterial membrane activity. b, Bar graphs showing the relative abundance (genus level) of in vitro en masse screening samples treated with peptides 1, 11, or their analogs as mode-of-action controls. Peptides 1 and 11 were each screened in triplicate in the screen. ‘HCl salt’ refers to peptides that had been converted from the trifluoroacetate counterion salt (obtained after preparative HPLC) to the hydrochloride salt. ‘Trifluoroacetate’ refers to a sample treated with sodium trifluoroacetate (1 mM). The peptide HCl salts and trifluoroacetate samples were used to establish that trifluoroacetate itself (present as counterions with the screened peptides) does not affect microbiota composition. c, Principal component analysis for in vitro en masse screening samples treated with peptides 1, 11, or their analogs as mode-of-action controls (n = 3 independent samples each for peptide 1 and peptide 11, n = 1 for the other treatments shown). As expected, peptides 1 and 11 promoted distinct microbiota remodeling activity that were clustered for the replicate treatments along with their corresponding HCl salt and enantiomeric peptide (which can self-assemble). On the other hand, the diastereomeric or N-methylated analogs (which cannot self-assemble) did not substantially affect microbiota growth, as demonstrated by their clustering with the vehicle-treated and trifluoroacetate control samples.
Extended Data Fig. 4 Design rationale and structures for mechanism-of-action control peptides.
In the absence of backbone N-methylation, as in peptide 11, the flat ring-shaped cyclic conformation favors peptide stacking and inter-subunit backbone hydrogen bonding, giving rise to tubular ensembles that can perturb transmembrane ion gradients to exert antimicrobial activities. Backbone N-methylation on each face of the macrocycle, as in peptide 30 and 33, creates a dual effect that prevents peptide self-assembly and membrane/antimicrobial activity. The modified ring structure not only lacks two amide hydrogen bonding sites but is also incapable of ring stacking and inter-subunit hydrogen bonding as a result of steric clashes by the N-methyl moieties. Therefore, peptides 30 and 33 are interesting control and mechanism of action probes because of their inability to self-assemble and exert membrane and antimicrobial activity, despite having identical amino acid sequence as peptide 11. For clarity, side chains are omitted from the molecular models shown. Likewise, switching the stereochemistry of one of the amino acid side chains yields a diastereomer of the parent peptide. Diastereomers, such as peptides 31, 32, and 35 lack the flat ring-shaped cyclic conformation that favors peptide stacking, thus diminishing the propensity for self-assembly. On the other hand, the alternating arrangement of D- and L-amino acids is present in enantiomers of the parent peptides, such as 34 and 36, producing the flat ring-shaped cyclic conformation that favors peptide stacking.
Extended Data Fig. 5 Daily oral administration of cyclic peptides c[wLwReQeR] and c[wLwKhShK] for 10 weeks showed no toxicity in vivo.
a,b, The body weights of the mice during the 10-week cyclic peptide treatments did not differ from vehicle controls. Data are shown as mean ± SD. c-f, The liver weights (c,d) and spleen weights (e,f) of peptide-treated animals did not significantly differ from vehicle controls. g-j, Plasma levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) of peptide-treated animals did not significantly differ from vehicle controls, indicating the cyclic peptides did not cause liver damage or injury in the mice. In panels c-i, the horizontal line shows the mean. In all panels, n = 8 mice for WD + vehicle group in the c[wLwReQeR] study; n = 7 mice for WD + c[wLwReQeR] group; n = 9 mice for WD + vehicle group in the c[wLwKhShK] study; n = 7 mice for WD + c[wLwKhShK] group.
Extended Data Fig. 6 Quantitation of peptide levels in feces of treated mice.
a, Representative LCMS selected ion traces used to generate standard curves for determining the concentration of peptides in extracted mouse feces. The traces correspond to ion 1181.8 ([M-H]1−) for c[wLwReQeR] and ion 368.2 ([M + 3 H]3+) for c[wLwKhShK] at the peptide concentrations shown. Standard curves were generated from two independent extractions. b, Standard curves used for quantitation of c[wLwReQeR] and c[wLwKhShK] concentrations in the feces of treated mice. Data are shown as mean ± SD of n = 2 independent replicates for each concentration. c, Measured quantities of feces (dry weight after lyophilization of fecal pellets) excreted by individually-housed WD-fed LDLr−/− mice over 24-h periods (n = 32). Data are shown as mean ± SD. The observed mean ± SD value was 432 ± 61 mg feces/day/mouse. d, Measured levels of fully intact peptides in the 5-wk fecal samples of treated LDLr−/− mice (n = 4 animals per group). Each circle represents the average of duplicate measurements from a single animal. Data are given as mean ± SD of the values for the 4 animals in each group. The observed fecal concentrations of the fully-intact peptides of 1.0 ± 0.1 nmol peptide/mg feces and 0.9 ± 0.2 nmol peptide/mg feces for c[wLwReQeR] and c[wLwKhShK], respectively, represent greater than 50% of the level that would be expected assuming all of the administered peptide was excreted in the feces (~1.7 nmol peptide/mg feces). The estimated maximum fecal peptide concentration that would be expected assuming all of the administered peptide was excreted in the feces was calculated from the known concentration of peptide administered in the drinking water (0.18 mM), the average daily volume of treated drinking water consumed by each mouse (4.5 mL), and the average daily amount of feces excreted by each mouse (432 mg).
Extended Data Fig. 7 Effects of peptide treatment on bacterial composition and richness over time.
a, Comparative effects of cyclic D,L-α-peptide-mediated remodeling of gut microbiota in LDLr−/− mice over the course of a 10-wk study. The time course analysis shows the number of genera in feces samples that were significantly changed in abundance relative to vehicle treatment at the timepoint indicated (n = 7 animals for all groups). The two peptides initially targeted different bacterial genera (out of 50 total genera that were significantly changed in abundance by treatment with the two peptides [adjusted p-value < 0.1, as determined by a hypergeometric test], only four genera were affected in common by both peptides after 2-wk peptide treatments). Over the course of the study, the remodeled gut microbiome community induced by peptide treatments became more similar (after 10-wk treatment, 15 bacterial genera were inhibited in common out of 39 total genera affected by the peptides). b, Effect of peptide treatment on richness (Chao1 index) of gut microbiota from WD-fed LDLr−/− mice (n = 7 animals for all groups). Samples were taken from feces after a 2-wk treatment period. The scatter plot is shown with mean ± SD. p values were determined by ANOVA. c, Peptide treatment shifted the ratio of Bacteroidetes to Firmicutes in feces of WD-fed mice toward that of the CHD-fed controls over the time periods shown. Scatter plots are shown with mean. p values were determined by two-tailed Student’s t-test. n = 5 per group. d, Principal-component analysis of genera abundance for CHD-fed mice and for WD-fed mice treated with peptides or vehicle (n = 6 animals each for CHD and WD groups; n = 8 animals each for c[wLwReQeR] and c[wLwKhShK] groups, data shown for 10-wk timepoint). e, Principal-component analysis of genera abundance for WD/vehicle and CHD/vehicle mice after 2-wk and 10-wk (n = 6 per group).
Extended Data Fig. 8 In vivo remodeling effects of cyclic D,L-α-peptides on the gut microbiota genera for which WD feeding caused significant changes in abundance compared to CHD.
a, Comparison of the bacterial genera observed to significantly differ (adusted p-value < 0.1, as determined by DESeq2 using a two-sided Wald test with adjustment for multiple comparisons using the Benjamini-Hochberg method) between WD-fed mice compared to CHD-fed mice from two independent animal studies (in both studies, n = 9 animals for CHD group and n = 8 animals for WD group). 18 genera were observed to differ significantly in common between the two independent studies. Eleven of the 18 genera became significantly more abundant and 7 genera became significantly less abundant after WD-feeding for two weeks. b, Heatmap showing the fold change of each of the 18 genera identified in panel (a) that differed in both independent in vivo studies. c, Plot of the abundance changes for the 18 genera identified in panel (a) for WD-feeding relative to CHD-feeding (red indicates more abundant in WD-fed animals and blue indicates less abundant in WD-fed animals). d, A heat map showing how oral peptide treatment affected the abundance of the 18 genera identified in panel (a). The negative correlation for c[wLwReQeR] against the bacteria that became more abundant in WD-feeding indicates that c[wLwReQeR] remodeled the gut microbiome by causing those genera to become less abundant. In contrast, c[wLwKhShK] treatment promoted the growth of the bacterial genera that became less abundant with WD-feeding, as indicated the positive correlation for c[wLwKhShK] against WD-less abundant genera. All microbiota samples in this figure were taken from 2-wk feces samples of LDLr−/− mice.
Extended Data Fig. 9 Cyclic peptide c[wLwReQeR] altered the gut microbiota transcriptome without drastically affecting the gut microbiota composition.
a, Peptide-mediated changes in gene expression could be grouped into three main clusters. Cluster 1 and Cluster 2 contained bacterial genes for which expression was increased or decreased, respectively, by peptide treatment. Cluster 3 contained genes that were altered by the WD compared the CHD, but not affected by peptide treatment. The Venn diagram reports the number of bacterial species having transcripts within each cluster. The majority of species had transcripts within each cluster, indicative of broad transcriptomic changes across the gut microbiome. b, Gene expression levels from each bacterial phylum for the three gene expression Clusters. RNA expression was increased from Bacteroidetes and decreased from Firmicutes in Cluster 1 compared to Cluster 2 or Cluster 3. c, Bacterial functions of the gut microbiome, as predicted by transcriptome analysis of feces samples from CHD fed mice. The values shown are the percentage of reads from the transcriptome analysis belonging to each category.
Extended Data Fig. 10 Comparison of gut microbiome gene expression levels for metabolic processes among the different treatment groups.
The graph shows gene expression levels of various metabolic processes for the CHD, WD, or c[wLwReQeR]-treated animals, as determined by RNA-Seq analysis of fecal samples taken after a 2-wk treatment period.
Supplementary information
Supplementary Information
Supplementary Note, Supplementary Figs. 1–4 and Supplementary Tables 1–8.
Rights and permissions
About this article
Cite this article
Chen, P.B., Black, A.S., Sobel, A.L. et al. Directed remodeling of the mouse gut microbiome inhibits the development of atherosclerosis. Nat Biotechnol 38, 1288–1297 (2020). https://doi.org/10.1038/s41587-020-0549-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-020-0549-5
This article is cited by
-
Chiral nanoparticle-remodeled gut microbiota alleviates neurodegeneration via the gut–brain axis
Nature Aging (2023)
-
Targeting the human gut microbiome with small-molecule inhibitors
Nature Reviews Chemistry (2023)
-
Nonfood Probiotic, Prebiotic, and Synbiotic Use Reduces All-Cause and Cardiovascular Mortality Risk in Older Adults: A Population-Based Cohort Study
The Journal of nutrition, health and aging (2023)
-
Artificial Intelligence for Risk Assessment on Primary Prevention of Coronary Artery Disease
Current Cardiovascular Risk Reports (2023)
-
Neutrophil activation and NETosis are the predominant drivers of airway inflammation in an OVA/CFA/LPS induced murine model
Respiratory Research (2022)