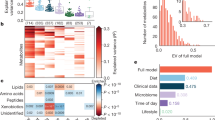
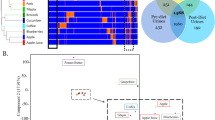
Abstract
Dietary assessment traditionally relies on self-reported data, which are often inaccurate and may result in erroneous diet–disease risk associations. We illustrate how urinary metabolic phenotyping can be used as an alternative approach to obtain information on dietary patterns. We used two multipass 24 h dietary recalls, obtained on two occasions on average 3 weeks apart, paired with two 24 h urine collections from 1,848 US individuals; 67 nutrients influenced the urinary metabotype (metabolic phenotype) of 46 structurally identified metabolites characterized by 1H NMR spectroscopy. We investigated the stability of each metabolite over time and showed that the urinary metabolic profile is more stable within individuals than reported dietary patterns. The 46 metabolites accurately predicted healthy and unhealthy dietary patterns in a free-living US cohort, and these predictions were replicated in an independent UK cohort. We mapped these metabolites into a host-microbial metabolic network to identify key pathways and functions related to diet. These data can be used in future studies to evaluate how this set of diet-derived, stable, measurable bioanalytical markers is associated with disease risk. This knowledge may give new insights into biological pathways that characterize the shift from a healthy to an unhealthy metabolic phenotype and hence indicate entry points for prevention and intervention strategies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data reported in this manuscript are tabulated in the main paper and in the Supplementary Tables. The NutriomeXplorer software contains all nutrient–metabolite associations and can be obtained from two separate public repositories (Figshare: https://doi.org/10.35092/yhjc.12181938, Box: https://imperialcollegelondon.box.com/s/f1in5lsnoh1hej5b8bvqr14tt7uoaq2v). The data that support the findings of this study are available from the corresponding authors upon request. Applications for access to the INTERMAP data can be made to the access committee (led by L.V.H.).
Code availability
The codes for executing the PLS, covariate-adjusted (O)PLS and simple orthogonal PLS/PLS-DA can be obtained from https://bitbucket.org/jmp111/capls/src/. The code for executing the STORM algorithm can be obtained from https://bitbucket.org/jmp111/storm/src. The codes for calculating the ACC, ICC and pICC can be obtained from https://bitbucket.org/jmp111/nutriome/src (this repository also contains a MATLAB version of the NutriomeXplorer). These can be executed in a MATLAB environment.
Change history
16 June 2021
A Correction to this paper has been published: https://doi.org/10.1038/s43016-021-00309-6
References
Rosell, M. S., Hellenius, M. L., de Faire, U. H. & Johansson, G. K. Associations between diet and the metabolic syndrome vary with the validity of dietary intake data. Am. J. Clin. Nutr. 78, 84–90 (2003).
Poslusna, K., Ruprich, J., de Vries, J. H., Jakubikova, M. & van’t Veer, P. Misreporting of energy and micronutrient intake estimated by food records and 24 hour recalls, control and adjustment methods in practice. Br. J. Nutr. 101(Suppl. 2), 73–85 (2009).
Freisling, H. et al. Dietary reporting errors on 24 h recalls and dietary questionnaires are associated with BMI across six European countries as evaluated with recovery biomarkers for protein and potassium intake. Br. J. Nutr. 107, 910–920 (2012).
Ioannidis, J. P. A. The challenge of reforming nutritional epidemiologic research. JAMA 320, 969–970 (2018).
Brennan, L. & Hu, F. B. Metabolomics-based dietary biomarkers in nutritional epidemiology—current status and future opportunities. Mol. Nutr. Food Res. 63, e1701064 (2019).
Guasch-Ferre, M., Bhupathiraju, S. N. & Hu, F. B. Use of metabolomics in improving assessment of dietary intake. Clin. Chem. 64, 82–98 (2018).
Ulaszewska, M. M. et al. Nutrimetabolomics: an integrative action for metabolomic analyses in human nutritional studies. Mol. Nutr. Food Res. 63, e1800384 (2019).
Price, N. D. et al. A wellness study of 108 individuals using personal, dense, dynamic data clouds. Nat. Biotechnol. 35, 747–756 (2017).
Holmes, E. et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400 (2008).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Nicholson, J. K. & Wilson, I. D. High-resolution proton magnetic-resonance spectroscopy of biological-fluids. Prog. Nucl. Magn. Reson. Spectrosc. 21, 449–501 (1989).
Gavaghan, C. L., Holmes, E., Lenz, E., Wilson, I. D. & Nicholson, J. K. An NMR-based metabonomic approach to investigate the biochemical consequences of genetic strain differences: application to the C57BL10J and Alpk:ApfCD mouse. FEBS Lett. 484, 169–174 (2000).
Stella, C. et al. Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 5, 2780–2788 (2006).
Nicholson, G. et al. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol. Syst. Biol. 7, 525 (2011).
Heinzmann, S. S. et al. Stability and robustness of human metabolic phenotypes in response to sequential food challenges. J. Proteome Res. 11, 643–655 (2012).
Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 (2012).
Garcia-Perez, I. et al. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol. 5, 184–195 (2017).
Gibbons, H. et al. Demonstration of the utility of biomarkers for dietary intake assessment; proline betaine as an example. Mol. Nutr. Food Res. 61, 1700037 (2017).
Scalbert, A. et al. The food metabolome: a window over dietary exposure. Am. J. Clin. Nutr. 99, 1286–1308 (2014).
Fenech, M. Nutrition and genome health. Forum Nutr. 60, 49–65 (2007).
Stamler, J. et al. INTERMAP: background, aims, design, methods, and descriptive statistics (nondietary). J. Hum. Hypertens. 17, 591–608 (2003).
Dennis, B. et al. INTERMAP: the dietary data—process and quality control. J. Hum. Hypertens. 17, 609–622 (2003).
Posma, J. M. et al. Integrated analytical and statistical two-dimensional spectroscopy strategy for metabolite identification: application to dietary biomarkers. Anal. Chem. 89, 3300–3309 (2017).
McLean, R. M. Measuring population sodium intake: a review of methods. Nutrients 6, 4651–4662 (2014).
Yi, S. S. & Kansagra, S. M. Associations of sodium intake with obesity, body mass index, waist circumference, and weight. Am. J. Prev. Med. 46, 53–55 (2014).
Elliott, P. et al. Urinary metabolic signatures of human adiposity. Sci. Transl. Med. 7, 285ra262 (2015).
Aburto, N. J. et al. Effect of lower sodium intake on health: systematic review and meta-analyses. BMJ 346, f1326 (2013).
Teague, C. et al. Ethyl glucoside in human urine following dietary exposure: detection by 1H NMR spectroscopy as a result of metabonomic screening of humans. Analyst 129, 259–264 (2004).
Dahl, H., Stephanson, N., Beck, O. & Helander, A. Comparison of urinary excretion characteristics of ethanol and ethyl glucuronide. J. Anal. Toxicol. 26, 201–204 (2002).
Svensson, B. G., Akesson, B., Nilsson, A. & Paulsson, K. Urinary-excretion of methylamines in men with varying intake of fish from the Baltic Sea. J. Toxicol. Environ. Health 41, 411–420 (1994).
Zhang, A. Q., Mitchell, S. C. & Smith, R. L. Dietary precursors of trimethylamine in man: a pilot study. Food Chem. Toxicol. 37, 515–520 (1999).
de Zwart, F. J. et al. Glycine betaine and glycine betaine analogues in common foods. Food Chem. 83, 197–204 (2003).
Heinzmann, S. S. et al. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. Am. J. Clin. Nutr. 92, 436–443 (2010).
Pujos-Guillot, E. et al. Mass spectrometry-based metabolomics for the discovery of biomarkers of fruit and vegetable intake: citrus fruit as a case study. J. Proteome Res. 12, 1645–1659 (2013).
Posma, J. M., Robinette, S. L., Holmes, E. & Nicholson, J. K. MetaboNetworks, an interactive Matlab-based toolbox for creating, customizing and exploring sub-networks from KEGG. Bioinformatics 30, 893–895 (2014).
Drewnowski, A. Defining nutrient density: development and validation of the nutrient rich foods index. J. Am. Coll. Nutr. 28, 421–426 (2009).
Mellen, P. B., Gao, S. K., Vitolins, M. Z. & Goff, D. C. Deteriorating dietary habits among adults with hypertension. Arch. Intern. Med. 168, 308–314 (2008).
Molitor, J. et al. Blood pressure differences associated with Optimal Macronutrient Intake Trial for Heart Health (OMNIHEART)-like diet compared with a typical American diet. Hypertension 64, 1198–1204 (2014).
Posma, J. M. et al. Optimized phenotypic biomarker discovery and confounder elimination via covariate-adjusted projection to latent structures from metabolic spectroscopy data. J. Proteome Res. 17, 1586–1595 (2018).
World Health Organization & Food and Agriculture Organization Diet, Nutrition and the Prevention of Chronic Diseases Technical Report Series 916 (World Health Organization, 2003).
Appel, L. J. et al. A clinical trial of the effects of dietary patterns on blood pressure. DASH Collaborative Research Group. N. Engl. J. Med. 336, 1117–1124 (1997).
Tasevska, N., Runswick, S. A. & Bingham, S. A. Urinary potassium is as reliable as urinary nitrogen for use as a recovery biomarker in dietary studies of free living individuals. J. Nutr. 136, 1334–1340 (2006).
Mente, A., Irvine, E. J., Honey, R. J. D. & Logan, A. G. Urinary potassium is a clinically useful test to detect a poor quality diet. J. Nutr. 139, 743–749 (2009).
Kesteloot, H. et al. Relation of urinary calcium and magnesium excretion to blood pressure: the International Study of Macro- and Micro-nutrients and Blood Pressure and the International Cooperative Study on Salt, other Factors, and Blood Pressure. Am. J. Epidemiol. 174, 44–51 (2011).
Garcia-Perez, I. et al. Urinary metabolic phenotyping the slc26a6 (chloride–oxalate exchanger) null mouse model. J. Proteome Res. 11, 4425–4435 (2012).
Midttun, O., Ulvik, A., Nygard, O. & Ueland, P. M. Performance of plasma trigonelline as a marker of coffee consumption in an epidemiologic setting. Am. J. Clin. Nutr. 107, 941–947 (2018).
Whitton, C. et al. National Diet and Nutrition Survey: UK food consumption and nutrient intakes from the first year of the rolling programme and comparisons with previous surveys. Br. J. Nutr. 106, 1899–1914 (2011).
Iwahori, T. et al. Six random specimens of daytime casual urine on different days are sufficient to estimate daily sodium/potassium ratio in comparison to 7-day 24-h urine collections. Hypertens. Res. 37, 765–771 (2014).
Wilson, T. et al. Spot and cumulative urine samples are suitable replacements for 24-hour urine collections for objective measures of dietary exposure in adults using metabolite biomarkers. J. Nutr. 149, 1692–1700 (2019).
Garcia-Perez, I. et al. An analytical pipeline for quantitative characterization of dietary intake: application to assess grape intake. J. Agric. Food Chem. 64, 2423–2431 (2016).
Dumas, M. E. et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP study. Anal. Chem. 78, 2199–2208 (2006).
Smith, L. M. et al. Large-scale human metabolic phenotyping and molecular epidemiological studies via 1H NMR spectroscopy of urine: investigation of borate preservation. Anal. Chem. 81, 4847–4856 (2009).
Keun, H. C. et al. Analytical reproducibility in 1H NMR-based metabonomic urinalysis. Chem. Res. Toxicol. 15, 1380–1386 (2002).
Nicholson, J. K. et al. Metabolic phenotyping in clinical and surgical environments. Nature 491, 384–392 (2012).
Garcia-Perez, I. et al. Dietary metabotype modelling predicts individual responses to dietary interventions. Nat. Food https://doi.org/10.1038/s43016-020-0092-z (2020).
Holmes, E. et al. Detection of urinary drug metabolite (Xenometabolome) signatures in molecular epidemiology studies via statistical total correlation (NMR) spectroscopy. Anal. Chem. 79, 2629–2640 (2007).
Dieterle, F., Ross, A., Schlotterbeck, G. & Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290 (2006).
Posma, J. M. et al. Subset Optimization by Reference Matching (STORM): an optimized statistical approach for recovery of metabolic biomarker structural information from 1H NMR spectra of biofluids. Anal. Chem. 84, 10694–10701 (2012).
Garcia-Perez, I. et al. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. (in the press).
Macdiarmid, J. & Blundell, J. Assessing dietary intake: who, what and why of under-reporting. Nutr. Res. Rev. 11, 231–253 (1998).
Maroni, B. J., Steinman, T. I. & Mitch, W. E. A method for estimating nitrogen intake of patients with chronic renal-failure. Kidney Int. 27, 58–65 (1985).
Mariotti, F., Tome, D. & Mirand, P. P. Converting nitrogen into protein—beyond 6.25 and Jones’ factors. Crit. Rev. Food Sci. Nutr. 48, 177–184 (2008).
Black, A. E. Critical evaluation of energy intake using the Goldberg cut-off for energy intake: basal metabolic rate. A practical guide to its calculation, use and limitations. Int. J. Obes. 24, 1119–1130 (2000).
Schofield, W. N. Predicting basal metabolic rate, new standards and review of previous work. Hum. Nutr. Clin. Nutr. 39(Suppl. 1), 5–41 (1985).
Fulgoni, V. L. III, Keast, D. R. & Drewnowski, A. Development and validation of the nutrient-rich foods index: a tool to measure nutritional quality of foods. J. Nutr. 139, 1549–1554 (2009).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005).
The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Hoffmann, C. et al. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS ONE 8, e66019 (2013).
Gaci, N., Borrel, G., Tottey, W., O’Toole, P. W. & Brugere, J. F. Archaea and the human gut: new beginning of an old story. World J. Gastroenterol. 20, 16062–16078 (2014).
Jaccard, P. Distribution de la flore alpine dans le bassin des Dranses et dans quelques régions voisines. Bull. Soc. Vaudoise Sci. Nat. 37, 241–272 (1901).
Newman, M. E. J. & Girvan, M. Finding and evaluating community structure in networks. Phys. Rev. E 69, 026113 (2004).
Guimera, R. & Nunes Amaral, L. A. Functional cartography of complex metabolic networks. Nature 433, 895–900 (2005).
Wang, Z. & Zhang, J. Z. In search of the biological significance of modular structures in protein networks. PLoS Comput. Biol. 3, 1011–1021 (2007).
Maslov, S. & Sneppen, K. Specificity and stability in topology of protein networks. Science 296, 910–913 (2002).
Acknowledgements
We thank the staff at local, national and international centres for collecting the INTERMAP data and samples. A partial listing of colleagues can be found in ref. 21. J.M.P. is supported by a Rutherford Fund Fellowship at Health Data Research (HDR) UK (MR/S004033/1). I.G.-P. is supported by a National Institute for Health Research (NIHR) fellowship (NIHR-CDF-2017-10-032). G.F. is an NIHR Senior Investigator. E.H. is supported by a Premier’s Science Fellowship (Western Australia). INTERMAP is supported by the US National Heart, Lung and Blood Institute (grant numbers R01-HL050490, R01-HL084228 and R01-HL135486), and received funding from the Chicago Health Research Foundation, and national agencies in Japan (grant number [A] 090357003) and the United Kingdom (project grant from the West Midlands National Health Service Research and Development, and grant number R2019EPH from the Chest, Heart and Stroke Association, Northern Ireland). Infrastructure support was provided by the NIHR Imperial Biomedical Research Centre (BRC). P.E. and E.H. acknowledge support from the UK Dementia Research Institute at Imperial College London, which receives funding from UK DRI Ltd funded by the Medical Research Council, the Alzheimer’s Society and Alzheimer’s Research UK. The funders had no role in study design.
Author information
Authors and Affiliations
Contributions
Conceptualization: J.M.P., P.E. and J.K.N. Methodology: J.M.P., E.H., P.E. and J.K.N. Software: J.M.P. Formal analysis: J.M.P. Investigation: J.M.P., I.G.-P. and G.A. Resources: L.V.H., M.D., J.S., E.H., P.E. and J.K.N. Writing—original draft: J.M.P., P.E. and J.K.N. Writing—review and editing: J.M.P., I.G.-P., G.F., G.S.A., Q.C., L.V.H., E.H., P.E. and J.K.N. Supervision: G.F., E.H., P.E. and J.K.N. Project administration: Q.C., L.V.H, M.D., J.S., E.H. and P.E. Funding acquisition: J.M.P., I.G.-P., L.V.H., M.D., J.S., E.H., P.E. and J.K.N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Representative partial 600 MHz 1H-NMR spectra of human urine showing selected assignments of some of the major metabolic signals.
Full 600 MHz 1H-NMR spectrum (mean of first visit data) is shown in 3 panels, top to bottom: δ 0.5–2.5 ppm, δ 2.5–4.5 ppm and δ 6.4–9.5 ppm. Metabolites found associated with intake of nutrients are labelled. Key: 1 – fatty acids (C5-C10), 2 – pantothenate, 3 – isoleucine, 4 – leucine, 5 – valine, 6 – 2-hydroxy-2-(4-methylcyclohex-3-en-1-yl)propoxyglucuronide, 7 – ethanol, 8 – ethyl glucuronide, 9 – 3-hydroxyisovalerate, 10 – alanine, 11 – unknown 1, 12 – acetate, 13 – phenylacetylglutamine, 14 – glutamine, 15 – O-acetylcarnitine, 16 – acetone, 17 – proline betaine, 18 – succinate, 19 – citrate, 20 – dimethylamine, 21 – S-methyl-cysteine-sulfoxide metabolite, 22 – N-acetyl-S-methyl-cysteine-sulfoxide, 23 – S-methyl-cysteine-sulfoxide metabolite, 24 – S-methyl-cysteine-sulfoxide, 25 – dimethylglycine, 26 – creatine, 27 – creatinine, 28 – N6,N6,N6-trimethyllysine, 29 – histidine, 30 – 1-methylhistidine, 31 – carnitine, 32 – taurine, 33 – trimethylamine-N-oxide, 34 – 4-hydroxyproline betaine, 35 – unknown 2, 36 – 4-hydroxyhippurate, 37 – hippurate, 38 – N-methylpyridinium, 39 – N-methylnicotinate, 40 – N-methylnicotinamide, 41 – N-methyl-2-pyridone-5-carboxamide, 42 – tyrosine, 43 – 3-hydroxymandelate, 44 – 2-furoylglycine, 45 – pseudouridine, 46 – formate.
Extended Data Fig. 2 Multicompartmental metabolic reaction network illustrating metabolic influence of 80 nutrients in the U.S. INTERMAP cohort (n = 1,848).
Grey nodes indicate the metabolites associated with one or more of the nutrients. Lines indicate reactions (mediated by Homo sapiens enzymes or by gut bacteria, see Methods). White nodes are intermediate metabolites connecting them (with 3 or more associated reactions) and white boxes are intermediate metabolites with two reactions. The background shading illustrates different types of metabolism based on closest affinity classification.
Extended Data Fig. 3 Schema of study design and exclusion criteria.
Scatter plots represent the expected/reported protein ratio (x-axis) and expected/reported energy ratio (y-axis) for both urine collections. The 95% confidence intervals (CI95) are indicated by the ellipses; red crosses indicate participants that mapped outside the CI95.
Supplementary information
Supplementary Information
Supplementary Figs. 1–17, Tables 1–7, Notes 1 and 2, Discussion, Methods and References.
Rights and permissions
About this article
Cite this article
Posma, J.M., Garcia-Perez, I., Frost, G. et al. Nutriome–metabolome relationships provide insights into dietary intake and metabolism. Nat Food 1, 426–436 (2020). https://doi.org/10.1038/s43016-020-0093-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43016-020-0093-y
This article is cited by
-
Towards objective measurements of habitual dietary intake patterns: comparing NMR metabolomics and food frequency questionnaire data in a population-based cohort
Nutrition Journal (2024)
-
Descriptive analysis of dietary (poly)phenol intake in the subcohort MAX from DCH-NG: “Diet, Cancer and Health—Next Generations cohort”
European Journal of Nutrition (2023)
-
Pathobionts in the tumour microbiota predict survival following resection for colorectal cancer
Microbiome (2023)
-
Metabolomic biomarkers of habitual B vitamin intakes unveil novel differentially methylated positions in the human epigenome
Clinical Epigenetics (2023)
-
Bioactives in the Food Supply: Effects on CVD Health
Current Atherosclerosis Reports (2022)