Abstract
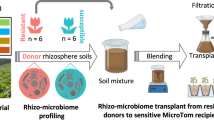
Tomato variety Hawaii 7996 is resistant to the soil-borne pathogen Ralstonia solanacearum, whereas the Moneymaker variety is susceptible to the pathogen. To evaluate whether plant-associated microorganisms have a role in disease resistance, we analyzed the rhizosphere microbiomes of both varieties in a mesocosm experiment. Microbiome structures differed between the two cultivars. Transplantation of rhizosphere microbiota from resistant plants suppressed disease symptoms in susceptible plants. Comparative analyses of rhizosphere metagenomes from resistant and susceptible plants enabled the identification and assembly of a flavobacterial genome that was far more abundant in the resistant plant rhizosphere microbiome than in that of the susceptible plant. We cultivated this flavobacterium, named TRM1, and found that it could suppress R. solanacearum-disease development in a susceptible plant in pot experiments. Our findings reveal a role for native microbiota in protecting plants from microbial pathogens, and our approach charts a path toward the development of probiotics to ameliorate plant diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
23 October 2018
In the version of this article initially published online, the citations to Supplementary Figures 12a and 12b were interchanged in section "TRM1 and disease resistance" and the last sentence of Results referred to the rhizosphere of Hawaii 7996; it should have referred to that of Moneymaker pretreated with TRM1-10. The error has been corrected in the print, PDF and HTML versions of this article.
References
Dangl, J.L., Horvath, D.M. & Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013).
Jones, J.D. & Dangl, J.L. The plant immune system. Nature 444, 323–329 (2006).
Hacquard, S., Spaepen, S., Garrido-Oter, R. & Schulze-Lefert, P. Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589 (2017).
Agrios, G. Plant Pathology (Elsevier Academic Press, Burlington, Massachusetts, USA, 2005).
Pieterse, C.M. et al. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375 (2014).
Lebeis, S.L. et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015).
Taiz, L., Zeiger, E., Møller, I.M. & Murphy, A. Plant Physiology and Development 761 (Sinauer Associates, Sunderland, Massachusetts, USA, 2015).
Hacquard, S. et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616 (2015).
Bulgarelli, D., Schlaeppi, K. & Spaepen, S. Ver Loren van Themaat, E. & Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 (2013).
Mendes, R., Garbeva, P., Raaijmakers, J.M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663 (2013).
Philippot, L., Raaijmakers, J.M., Lemanceau, P. & van der Putten, W.H. Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799 (2013).
Berg, G., Rybakova, D., Grube, M. & Köberl, M. The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002 (2016).
Castrillo, G. et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature 543, 513–518 (2017).
Lundberg, D.S. et al. Defining the core Arabidopsis thaliana root microbiome. Nature 488, 86–90 (2012).
Bulgarelli, D. et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95 (2012).
Bulgarelli, D. et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17, 392–403 (2015).
Müller, D.B., Vogel, C., Bai, Y. & Vorholt, J.A. The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234 (2016).
Busby, P.E. et al. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 15, e2001793 (2017).
Leach, J.E., Triplett, L.R., Argueso, C.T. & Trivedi, P. Communication in the Phytobiome. Cell 169, 587–596 (2017).
Walters, W.A. et al. Large-scale replicated field study of maize rhizosphere identifies heritable microbes. Proc. Natl. Acad. Sci. USA 115, 7368–7373 (2018).
Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Hayward, A.C. Biology and epidemiology of bacterial wilt caused by pseudomonas solanacearum. Annu. Rev. Phytopathol. 29, 65–87 (1991).
Mansfield, J. et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629 (2012).
Elphinstone, J.G. Bacterial wilt disease and the Ralstonia solanacearum species complex 9–28 (APS Press, St. Paul, Minnesota, USA, 2005).
Huet, G. Breeding for resistances to Ralstonia solanacearum. Front. Plant Sci. 5, 715 (2014).
Wang, J.F. et al. Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain-specific locus. Mol. Plant Microbe Interact. 13, 6–13 (2000).
Wu, J. et al. Loss of glutamate dehydrogenase in Ralstonia solanacearum alters dehydrogenase activity, extracellular polysaccharide production and bacterial virulence. Physiol. Mol. Plant Pathol. 90, 57–64 (2015).
Galperin, M.Y., Makarova, K.S., Wolf, Y.I. & Koonin, E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269 (2015).
Hu, L. et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738 (2018).
Albertsen, M. et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat. Biotechnol. 31, 533–538 (2013).
Kim, B.Y. et al. Flavobacterium daejeonense sp. nov. and Flavobacterium suncheonense sp. nov., isolated from greenhouse soils in Korea. Int. J. Syst. Evol. Microbiol. 56, 1645–1649 (2006).
Banning, E.C., Casciotti, K.L. & Kujawinski, E.B. Novel strains isolated from a coastal aquifer suggest a predatory role for flavobacteria. FEMS Microbiol. Ecol. 73, 254–270 (2010).
Naseem, M., Kaltdorf, M. & Dandekar, T. The nexus between growth and defense signaling: auxin and cytokinin modulate plant immune response pathways. J. Exp. Bot. 66, 4885–4896 (2015).
Aznar, A. & Dellagi, A. New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J. Exp. Bot. 66, 3001–3010 (2015).
Matilla, M.A. et al. Pseudomonas putida KT2440 causes induced systemic resistance and changes in Arabidopsis root exudation. Environ. Microbiol. Rep. 2, 381–388 (2010).
Planchamp, C., Glauser, G. & Mauch-Mani, B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 5, 719 (2015).
Wang, J.F. et al. Identification of major QTLs associated with stable resistance of tomato cultivar 'Hawaii 7996' to Ralstonia solanacearum. Euphytica 190, 241–252 (2013).
Mendes, R. et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011).
Weller, D.M., Raaijmakers, J.M., Gardener, B.B. & Thomashow, L.S. Microbial populations responsible for specific soil suppressiveness to plant pathogens. Annu. Rev. Phytopathol. 40, 309–348 (2002).
Cha, J.Y. et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. 10, 119–129 (2016).
Sangwan, N., Xia, F. & Gilbert, J.A. Recovering complete and draft population genomes from metagenome datasets. Microbiome 4, 8 (2016).
Buchan, A., LeCleir, G.R., Gulvik, C.A. & González, J.M. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698 (2014).
Valls, M., Genin, S. & Boucher, C. Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2, e82 (2006).
Genin, S. & Denny, T.P. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50, 67–89 (2012).
Meng, F., Babujee, L., Jacobs, J.M. & Allen, C. Comparative transcriptome analysis reveals cool virulence factors of Ralstonia solanacearum race 3 biovar 2. PLoS One 10, e0139090 (2015).
Wei, Z. et al. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat. Commun. 6, 8413 (2015).
Bai, Y. et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369 (2015).
Kim, J.F. et al. Genome sequence of the polymyxin-producing plant-probiotic rhizobacterium Paenibacillus polymyxa E681. J. Bacteriol. 192, 6103–6104 (2010).
Bordenstein, S.R. & Theis, K.R. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226 (2015).
Arnold, C. The hologenome: A new view of evolution. New Sci. 2899, 30–34 (2013).
Thoquet, P. et al. Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii 7996. Mol. Plant Microbe Interact. 9, 826–836 (1996).
Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Lan, Y., Wang, Q., Cole, J.R. & Rosen, G.L. Using the RDP classifier to predict taxonomic novelty and reduce the search space for finding novel organisms. PLoS One 7, e32491 (2012).
Jonsson, V., Österlund, T., Nerman, O. & Kristiansson, E. Statistical evaluation of methods for identification of differentially abundant genes in comparative metagenomics. BMC Genomics 17, 78 (2016).
Heo, J. et al. Gut microbiota modulated by probiotics and Garcinia cambogia extract correlate with weight gain and adipocyte sizes in high fat-fed mice. Sci. Rep. 6, 33566 (2016).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Zhu, W., Lomsadze, A. & Borodovsky, M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 38, e132 (2010).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Mavromatis, K. et al. The DOE-JGI standard operating procedure for the annotations of microbial genomes. Stand. Genomic Sci. 1, 63–67 (2009).
Kong, H.G. et al. Induction of the viable but nonculturable state of Ralstonia solanacearum by low temperature in the soil microcosm and its resuscitation by catalase. PLoS One 9, e109792 (2014).
Roberts, D.P., Denny, T.P. & Schell, M.A. Cloning of the egl gene of Pseudomonas solanacearum and analysis of its role in phytopathogenicity. J. Bacteriol. 170, 1445–1451 (1988).
Reasoner, D.J. & Geldreich, E.E. A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7 (1985).
Lamprecht, A.L. et al. GeneFisher-P: variations of GeneFisher as processes in Bio-jETI. BMC Bioinformatics 9 (Suppl. 4), S13 (2008).
Dupont, C.L. et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 6, 1186–1199 (2012).
Bowers, R.M. et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731 (2017).
Park, B.H., Karpinets, T.V., Syed, M.H., Leuze, M.R. & Uberbacher, E.C. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20, 1574–1584 (2010).
Saier, M.H. Jr., Reddy, V.S., Tamang, D.G. & Västermark, A. The transporter classification database. Nucleic Acids Res. 42, D251–D258 (2014).
Schaad, N.W., Jones, J.B. & Chun, W. Laboratory guide for identification of plant Pathogenic bacteria (American Phytopathological Society Press, St. Paul, USA, 2001).
Opina, N. et al. A novel method for development of species and strain specific DNA probes and PCR primers for identifying Burkholderia solanacearum (formerly Pseudomonas solanacearum). Asia Pac. J. Mol. Biol. Biotechnol. 5, 19–30 (1997).
Acknowledgements
We would like to thank members of the laboratories of J.F.K. and S.-W.L., including B.K. Kim, K.Y. Baek, T.-H. Kang, S. Kim, H.G. Lee, S.Y. Lee, G.J. Son, S. Yoo and H. Yu, as well as KRIBB-KOBIC and NABIC, for technical support, and Y.-S. Bahn, D. Choi, S.-Y. Kwon, I. Lee, W.-J. Lee and H.-S. Pai for helpful comments and suggestions. This study was financially supported by the Strategic Initiative for Microbiomes in Agriculture and Food (914001-4 to J.F.K. and 914006-4 to J.Y.S.), the Cooperative Research Program for Agricultural Science & Technology Development (PJ01093901 to S.-W.L.), the National Research Foundation (NRF-2014M3C9A33068822 and NRF-2011-0017670 to J.F.K.), and the Next-Generation BioGreen 21 Program (PJ008201 to S.-W.L.) of the Republic of Korea. Publication was supported in part by the Brain Korea 21 PLUS program, and M.-J.K., S.-K.K. and J.L. are fellowship awardees of the program.
Author information
Authors and Affiliations
Contributions
J.F.K. and S.-W.L. conceived, organized and supervised the project. J.F.K., S.-W.L., M.M.L. and E.M.R. interpreted the results and prepared the manuscript. People in the laboratory of S.-W.L. performed the plant experiments; those in J.F.K.'s lab. analyzed the metagenomic data. M.-J.K. worked on the metagenome analysis, reconstructed TRG1, and drafted the microbiome results. H.J.L. contributed to setting up the plant experiment. H.G.K. and K.C. extracted the metagenomic DNA and analyzed the pyrosequencing data. J.Y.S. carried out the comparative analysis on field and pot experiments. M.S. and H.K. performed the statistical analysis for community structures. S.Y.C. and E.J.J. performed the transplant experiment as well as the isolation and phenotypic characterization of flavobacteria. K.C. and P.A.L. tested the influence of root exudates on bacterial growth. J.Y.S. annotated TRG1. M.-J.K., S.-K.K., and J.L. analyzed the genome information. J.L. and M.-J.K. isolated and characterized TRM1. P.A.L., J.L., and K.C. tested its effect as well as those of other flavobacteria on disease progress. Finally, S.-K.K., P.A.L., and N.R. performed the qPCR analysis. K.C., P.A.L., H.P., and N.R. enumerated cultured bacteria. J.F.K. composed the main text. J.F.K., S.-W.L., and M.-J.K. edited the manuscript. All of the authors read and approved the final version of the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Plot design for metagenomic analysis of the plant rhizosphere.
(a) A foreground view of the plastic house at the Dong-A University Agricultural Experimental Station, where plants were cultivated. (b) Tomato plants uprooted during the prolific growth and first flowering stage. (c) Mesocosm experimental plots to grow tomato plants and Korean cabbage. Each colored square represents an individual plant. 1, plants used for the first rhizosphere sampling during the active growth and first flowering stage; 2, plants used for the second rhizosphere sampling during the fruiting and senescence stage.
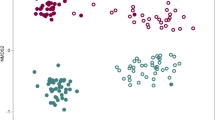
Supplementary Figure 2 Structures of the rhizosphere and bulk soil microbiotas based on 16S rDNA-amplicon pyrosequencing.
(a) Comparison of the rarefaction curves of the samples. Species richness was calculated at the 3% dissimilarity level. First, active growth and first flowering stage; second, fruiting and senescence stage. (b) Comparison of the diversity indices of the samples at the 3% dissimilarity level. ACE and Chao indicate the richness diversity indices. Inverse Simpson and Shannon indicate the evenness diversity indices. (c) Principal coordinate analysis. Principal coordinate analysis (PCoA) was performed with the representative sequences at the 3% dissimilarity level: bulk soil 5,134/3,981 (active growth/senescence), Korean cabbage 2,791/2,274, Moneymaker 2,265/2,859, and Hawaii 7996 2,425/1,952. PCoA plot shows that there are distinct differences between the microbial communities of bulk and planted soils, and between the first and second sampling stages. (d) Taxonomic comparison at the class level using 16S rDNA reads from amplicon sequencing. Circles indicate the bacterial classes with ≥ 0.1% relative abundance in both samples. Colored circles indicate ≥ 15% abundant taxa with ≥ 0.5% relative abundance in Hawaii 7996 or Moneymaker, as compared with each other. (e) Taxonomic comparison at the class level using the pyrosequencing reads of 16S rDNA amplicons. Taxonomic comparison was conducted with the average relative abundance of 10 subsets, which were randomly sub-sampled from 454 pyrosequencing data of Hawaii 7996 and Moneymaker. Taxa which relative abundance was higher than 0.5% in Hawaii 7996 or Moneymaker were shown as a bar graph. The bar plots indicate the average relative abundance of each sample in 10 subsets and the error bar indicates the standard error of the 10 subsets. Relative abundances of Alphaproteobacteria and Flavobacteriia are shown in the left Y-axis and relative abundances of the other taxa are shown in the right Y-axis. Statistical analysis was performed using one-way ANOVA, Kruskal-Wallis test in SPSS program. A total of 79 bacterial classes were analyzed for the statistical test. Among them, 13 bacterial classes had significant p-values (*, p < 0.05; **, p < 0.005; ***, p < 0.0005): Flavobacteriia 0.0002, Betaproteobacteria 0.0002, Gammaproteobacteria 0.0009, unclassified phylum 0.0015, Anaerolineae 0.0494, unclassified Proteobacteria 0.0002, Acidobacteria Gp4 0.0041, Bacteroidetes incertae sedis 0.0052, Cyanobacteria 0.0002, Acidobacteria Gp6 0.0233, Saccharibacteria genera incertae sedis 0.0002, Cytophagia 0.0041, Chloroflexia 0.0009. The dot plots in the below the bar plot were generated with same data used in the bar plot. Each circle indicates the distribution of relative abundance for each taxon. (f) Taxonomic comparison at the family level. Circles indicate the bacterial families with ≥ 0.1% relative abundance in both samples. Colored circles are the families that have the relative abundant of ≥ 0.5% in either of the samples and abundance difference of ≥ 20% when compared with each other. (g) Comparison of the relative abundances of taxa between Hawaii 7996 and bulk soil in the mesocosm and pot experiments. Bacterial abundance values in Hawaii 7996 relative to bulk soil were log-transformed. The plot shows bacterial families with ≥ 0.1% relative abundance and average relative abundance values in Hawaii 7996 were indicated by circle size. A total of 67 bacterial families are shown in the figure. The families with ≰ 0.05 FDR-adjusted p-value were highlighted by red color. (h) Comparison of the relative abundances of taxa between Moneymaker and bulk soil in the mesocosm and pot experiments. Bacterial abundance values in Moneymaker relative to bulk soil were log-transformed. The plot shows bacterial families with ≥ 0.1% relative abundance and average relative abundance values in Moneymaker were indicated by circle size. A total of 67 bacterial families are shown in the figure. The families with ≰ 0.05 FDR-adjusted p-value were highlighted by blue color.
Supplementary Figure 3 Comparison of the microbiota structures using the second batch of sequences from field experiment.
To confirm the results from the first community analysis, the 16S rRNA genes were amplified from additional samples before undergoing pyrosequencing. (a) Taxonomic comparison at the phylum level. First, active growth and first flowering stage; second, fruiting and senescence stage. (b) Taxonomic comparison at the class level. (c) Taxonomic comparison at the family level. (d) Comparison of the rarefaction curves of the samples. Species richness was calculated at the 3% dissimilarity level. (e) Comparison of the diversity indices of the samples at the 3% dissimilarity level. ACE and Chao indicate the richness diversity indices. Inverse Simpson and Shannon indicate the evenness diversity indices. (f) Correlation of the microbial community at the class level between resulted from the first sequencing data used in Fig. 1a and Supplementary Figs. 2a-f, and the second sequencing data used in Supplementary Fig. 3. A total of 80 bacterial classes are shown in the figure. Each circle represents the bacterial taxon.
Supplementary Figure 4 Comparison of the microbiota structures using the third batch of sequences from pot experiment.
To confirm the results from the first community analysis, Hawaii 7996 and Moneymaker were grown in pots with the field soil from the mesocosm experiment. After 45 days, metagenomic DNA was extracted from tomato rhizosphere at the active growth and first flowering stage. Amplification and pyrosequencing of 16S rRNA gene were performed separately using the metagenomic DNA. (a) Taxonomic comparison at the phylum level. First, active growth and first flowering stage; second, fruiting and senescence stage. (b) Taxonomic comparison at the class level. (c) Taxonomic comparison at the family level. (d) Comparison of the rarefaction curves of the samples. Species richness was calculated at the 3% dissimilarity level. (e) Comparison of the diversity indices of the samples at the 3% dissimilarity level. ACE and Chao indicate the richness diversity indices. Inverse Simpson and Shannon indicate the evenness diversity indices. (f) Correlation of the microbial community at the class level between resulted from the first sequencing data used in Fig. 1 and Supplementary Figs. 2a-f, and the third sequencing data used in Supplementary Fig. 3. A total of 80 bacterial classes are shown in the figure. Each circle represents the bacterial taxon.
Supplementary Figure 5 Microbiota structures in the rhizospheres of Hawaii 7996 and Moneymaker based on whole-metagenome sequencing and comparison of the metagenomic gene contents.
(a) Relative abundances of the microbial domains in whole-metagenomic data of the tomato rhizospheres. Relative abundances of each domain were calculated using the small-subunit rDNA reads extracted from the whole-metagenome sequences by read alignment against the Silva database. (b) Taxonomic comparison at the class level using whole metagenome-derived 16S rDNA reads. Circles indicate the bacterial classes with ≥ 0.1% relative abundance in both samples. Colored circles indicate ≥ 15% abundant taxa with ≥ 0.5% relative abundance in Hawaii 7996 or Moneymaker, as compared with each other. (c) Correlation between the bacterial community structures derived from the datasets of 16S rDNA amplicon pyrosequencing and those from Illumina whole-metagenome sequencing. The values were calculated as the ratio between relative abundance of the bacterial classes in Hawaii 7996 and relative abundance of the bacterial classes in Moneymaker. (d) Taxonomic distribution of CDSs specific to Hawaii 7996 or Moneymaker. Among 4,180,636 CDSs in the rhizosphere of Hawaii 7996, 203,204 were detected only in Hawaii 7996 and among 4,165,753 CDSs in the rhizosphere of Moneymaker, 188,321 were detected only in Moneymaker. Among these, 73,031 CDSs in Hawaii 7996 and 67,808 CDSs in Moneymaker were taxonomically assigned to bacterial or archaeal classes. The abundance of each CDS was calculated using RPK, and taxonomic assignment of each CDS was conducted with BLASTP against the GenBank NR database. Taxonomic abundance was calculated using the sum of the RPKs of each CDS. (e) Difference in the relative abundance of cultivar-specific COG-assigned CDSs. Red bars indicate the COGs more abundant in Hawaii 7996 and blue bars indicate the COGs more abundant in Moneymaker. Among the cultivar-specific CDSs, 36,686 in Hawaii 7996 and 34,359 in Moneymaker were assigned to the COG categories. COG categories: A, RNA processing and modification; B, chromatin structure and dynamics; C, energy production and conversion; D, cell cycle control, cell division, chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination, and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; Q, secondary metabolites biosynthesis, transport, and catabolism; R, general function prediction only; S, function unknown; T, signal transduction mechanisms; U, intracellular trafficking and secretion; V, defense mechanisms; W, extracellular structures; Z, cytoskeleton. (f) Difference in the relative abundance of COG-assigned CDSs, which are more than twice abundant in either cultivar. Red bars indicate the COGs more abundant in Hawaii 7996 and blue bars indicate the COGs more abundant in Moneymaker. Among the CDSs that are abundant more than twice in each cultivar, 265,838 in Hawaii 7996 and 249,907 in Moneymaker were assigned to the COG categories.
Supplementary Figure 6 Comparison of the rhizospheric gene contents of Hawaii 7996 and Moneymaker.
(a and b) Taxonomic distribution of CDSs occurring more than twice in Hawaii 7996 or Moneymaker, when compared with each other. 817,106 and 762,818 CDSs were abundant more than twice in Hawaii 7996 or Moneymaker, respectively. Among them, 454,236 and 432,248 CDSs were taxonomically assigned to the bacterial or archaeal classes. (c) Relative abundance of COG-assigned CDSs in the rhizosphere of Hawaii 7996 and Moneymaker. 1,570,962 and 1,568,636 CDSs were assigned to the COG category in Hawaii 7996 and Moneymaker, respectively. Functions of individual COG categories are described in Online Methods.
Supplementary Figure 7 Disease progress of bacterial wilt on the transplanted tomato cultivars.
Distributions of the disease indices of transplanted tomato cultivars were presented by box plots. Four box plots show the disease indices at four different time points (6, 8, 10, and 12 days) from Fig. 2b. The bottom and top edges of the box indicate the value corresponds to lowest 25 % and the value corresponds to highest 75%, respectively. The bottom and top error whiskers indicate the minimum and maximum values, respectively. Whiskers are the range of the data extended to the extreme data point of 1.5× interquartile range from the edge of the box. Bold horizontal line in the box indicates the median value of the disease indices. Each data point represents the mean disease index for three independent experiments (n = 18 in total for susceptible plants, n = 20 in total for resistant plants for each treatment). Open circles indicate the outlier values in each data set. Statistical significances between the transplanted tomato cultivars were evaluated by Wilcoxon signed ranks test using IBM SPSS 23 software. S/S, Moneymaker in Moneymaker-grown soil; S/R, Moneymaker in Hawaii 7996-grown soil; R/S, Hawaii 7996 in Moneymaker-grown soil; R/R, Hawaii 7996 in Hawaii 7996-grown soil. *, p < 0.05; **, p < 0.005. p values: Day 6, S/S vs. R/R (0.019), S/S vs. R/S (0.018); Day 8, S/S vs. R/R (0.001), S/S vs. R/S (0.001), S/R vs. R/R (0.015), S/R vs. R/S (0.031); Day 10, S/S vs. S/R (0.044), S/S vs. R/R (0.001), S/S vs. R/S (0.002), S/R vs. R/R (0.027); Day 12, S/S vs. R/R (0.001), S/S vs. (R/S 0.003).
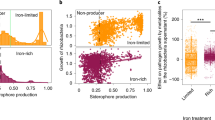
Supplementary Figure 8 Effects of the root exudates on the growth of Ralstonia solanacearum and isolation of flavobacterial strains from the rhizosphere to screen for involvement in disease resistance.
Growth of R. solanacearum SL341 in M9 broth supplemented with glucose (2%) and/or root exudates of tomato cultivars, Hawaii 7996 and Moneymaker. (a) Growth rates of SL341 in M9 broth with glucose (M9G) shake-cultured at 120 rpm. Red color, SL341 in M9G supplemented with root exudate of Hawaii 7996; blue, SL341 in M9G with root exudate of Moneymaker; green, SL341 in M9G with aqueous extract of bulk soil; black, SL341 in M9G broth. (b) Growth of SL341 in M9 broth without glucose (M9) shake-cultured at 120 rpm. Red color, SL341 in M9 with root exudate of Hawaii 7996; blue, SL341 in M9 with root exudate of Moneymaker; green, SL341 in M9 with aqueous extract of bulk soil; black, SL341 in M9. Vertical bars indicate the standard deviations from 3 replications. (c) A scheme to isolate flavobacteria from the tomato rhizosphere. Bacteria from the tomato rhizosphere and bulk soil were grown on TSA or R2A medium, and compared to select apparently unique bacterial colonies. A total of 477 colonies were PCR-amplified to select 32 flavobacterial isolates. (d) Antifungal activities of selected Flavobacterium strains (colonies on the right) against Phytophthora infestans (white mycelia on the left). The antifungal activities of bacterial strains have been repeated at least three times independently.
Supplementary Figure 9 Comparative genomic analysis of TRG1.
(a) Number of genomes with < 1× genome coverage (black) in the whole-metagenome dataset of this study and number of genomes with ≥ 1× genome coverage (gray). Completely sequenced bacterial genomes were downloaded from the NCBI Genome database and only one genome of each species was selected for a reference-guided analysis. Genome selection criteria were described in Online Methods. Among the total reads, 6.66% and 6.46% of the reads aligned to the reference genomes for Hawaii 7996 and Moneymaker, respectively. Among the 168 genomes, there was no flavobacterial genome. (b) Shared or unique genes among TRM1, RCH33, DSM 17708, and other flavobacterial strains (all marine) in the same clade as TRM1 (indicated in a bracket in Fig. 3c). Among 229 flavobacterial genomes including draft sequences, only those of TRG1, RCH33, and DSM 17708 were detected in the tomato rhizosphere with ≥ 1× genome coverage. RCH33 was isolated from the tomato rhizosphere in this study. DSM 17708 was isolated from a greenhouse soil in Korea. (c) Organization of the 128 rhizospheric strain-specific gene clusters in TRG1. Red and green colored arrows indicate the rhizospheric strain-specific genes and gray arrows are genes that are not rhizospheric strain-specific. Green genes encode carbohydrate-active enzymes. (d) Comparison of COG distribution. Relative abundances were calculated using the total number of COG-assigned CDSs. (e) Gene organization of contig 24. This contig contains a number of genes encoding glycosyl modification enzymes including glycosyl transferases, glycoside hydrolases, and acetyltransferases in a large gene cluster. More than half of the genes in this contig were not found in the Flavobacteriaceae genomes compared in (b) above.
Supplementary Figure 10 Cultivation and isolation of TRM1.
(a) Confirmation of the existence of the targeted bacterium in the rhizosphere sample by PCR. 1–3, TRM1-specific primer sets (1, rpoA; 2, recA; 3, secY); +, 16S rRNA universal primer set (27F and 1492R). (b) Confirmation of the presence of the targeted bacterium using genomic DNA of total bacteria from the enrichment broth culture by PCR. For each medium, TRM1-specific primers for rpoA, recA, and secY, and 16S rRNA universal primers (27F and 1492R), respectively, were used from left to right. (c) TRM1 colonies grown on agar plates containing 1/10 MB, 2% NaCl, and 30 μg/ml kanamycin. (d) PCR detection of 22 TRM1 isolates using specific primer sets. For each isolate, TRM1-specific primers for rpoA, recA, and secY, and 16S rRNA universal primers (27F and 1492R), respectively, were used from left to right. M, DNA ladder; +, Genomic DNA extracted from the enrichment culture. (e) TRM1-10 colonies grown on a marine agar plate.
Supplementary Figure 11 Disease progress of bacterial wilt on Moneymaker treated with TRM1-10.
Distributions of the disease indices of Moneymaker plants inoculated with R. solanacearum SL341 alone (labeled C in the X-axis) or Flavobacteriaceae sp. TRM1-10 at different concentrations (CFU/g soil) and SL341 were presented by box plots. Four box plots show the disease indices at four different time points (4, 6, 8, and 10 days) from Fig. 5c. The bottom and top edges of the box indicate the value corresponds to lowest 25% and the value corresponds to highest 75%, respectively. Whiskers are the range of the data extended to the extreme data point of 1.5× interquartile range from the edge of the box. The bottom and top error whiskers indicate the minimum and maximum values, respectively. Bold horizontal line in the box indicates the median value (n = 30 in total per treatment) of the disease indices. Open circles indicate the outlier values in each data set. Statistical significances between the treatments were evaluated by Wilcoxon signed ranks test using IBM SPSS 23 software. *, p < 0.05. p values: Day 8, control vs. 2×108 cells (0.026); Day 10, control vs. 2×108 cells (0.006).
Supplementary Figure 12 Effects of TRM1-10, three Flavobacterium strains, and P. putida KT2440 on the progress of bacterial wilt in Moneymaker.
Disease progress of bacterial wilt on Moneymaker treated with TRM1-10, F. aquidurense RC62, F. daejeonense RCH33, Flavobacterium sp. TCH3-2, or P. putida KT2440 using either (a) sterile commercial horticultural nursery soil or (b) non-sterile commercial horticultural nursery soil mixed with non-sterile natural field soil. One week prior to the inoculation of R. solanacearum SL341, tomato plants were treated with TRM1-10, each of the three Flavobacterium strains, or P. putida KT2440. Repeated measures ANOVA displayed a significant difference between TRM1-10 treatment and control treatment in sterile nursery soil (*, p = 0.01474) and in field soil mixed with non-sterile nursery soil (*, p = 0.01731). Treatment of each of the Flavobacterium strains was not significantly different from the non-treatment control for bacterial wilt suppression. Significant interaction between TRM1-10 treatment and experimental days were noticed for both cases: sterile nursery soil (p < 2.2e−16) and field soil mixed with non-sterile nursery soil (p = 4.013e−5). Treatment of P. putida KT2440 showed a significant difference as compared to the non-treated control only in sterile nursery soil (p = 0.0003345). Each vertical bar represents the standard error of the means from three replicates (each replicate with 10 plants, n = 30 in total for each treatment). Disease index: 0, no disease symptoms; 1, 1-25% of leaves wilted; 2, 26-50% of leaves wilted; 3, 51-75% of leaves wilted; 4, 76-100% of leaves wilted.
Supplementary Figure 13 Population dynamics and disease progress of TRM1-10 (TRM1) and R. solanacearum SL341 (Rsol) in tomato plants.
(a) Pearson correlation showing a strong positive correlation between colony count and qPCR throughout all experimental periods for TRM1-10 (left) and R. solanacearum SL341 (right) populations (n = 47 for TRM1, n = 28 for Rsol). (b) Population changes of TRM1-10 and R. solanacearum SL341 in bulk soil over time. Two-sample, two-sided t-tests were used to compare bacterial populations between samples (n = 5). The population of TRM1-10 remained stable in the bulk soil of Hawaii 7996. On the other hand, the population of TRM1-10 in bulk soil remarkably decreased in Moneymaker to almost undetectable levels. TRM1-10 populations differed between the two cultivars and were significantly higher in Hawaii 7996 than Moneymaker on D7 (p = 0.034), D10 (p = 3.25e−5), and D14 (p = 0.004). In Moneymaker, TRM1-10 showed significantly higher populations across all sample days, i.e. D7 (p = 0.039), D10 (p = 3.90e−4) and D14 (p = 0.003), treated with TRM1-10 and SL341 than TRM1-10 alone. The populations of SL341 decreased significantly (p = 5.32e−4) from D7 to D10 in TRM1+Rsol, but no significant difference was observed between D10 and D14 (p = 0.689). The populations of SL341 in Rsol alone and TRM1+Rsol showed no significant difference (p = 0.601) on D10; it was significantly higher in Rsol alone on D14 (p = 0.013). Each vertical bar represents the standard deviation of the mean from 5 plants. Asterisks indicate significant difference between treatments (*, p < 0.05; **, p < 0.01; ***, p < 0.001). (c) Bacterial wilt suppression by TRM1-10 and in planta population of SL341 in Moneymaker. Repeated measures ANOVA displayed a significant suppression of bacterial wilt in TRM1+Rsol than Rsol alone (*, p = 0.02635), a significant difference between experimental periods (p = 9.389e−9), and a significant interaction between treatments and experimental days (p = 2.572e−9). Each vertical bar represents the standard deviation of the mean from 14 plants remained until experimental period D14.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 (PDF 7371 kb)
Supplementary Tables
Supplementary Tables 1–18 (PDF 17906 kb)
Rights and permissions
About this article
Cite this article
Kwak, MJ., Kong, H., Choi, K. et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36, 1100–1109 (2018). https://doi.org/10.1038/nbt.4232
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4232
This article is cited by
-
Selective regulation of endophytic bacteria and gene expression in soybean by water-soluble humic materials
Environmental Microbiome (2024)
-
Analysis of endophytic bacterial diversity in seeds of different genotypes of cotton and the suppression of Verticillium wilt pathogen infection by a synthetic microbial community
BMC Plant Biology (2024)
-
The beneficial endophytic microbes enhanced tobacco defense system to resist bacterial wilt disease
Chemical and Biological Technologies in Agriculture (2024)
-
Microbiome homeostasis on rice leaves is regulated by a precursor molecule of lignin biosynthesis
Nature Communications (2024)
-
Leaf microbiome dysbiosis triggered by T2SS-dependent enzyme secretion from opportunistic Xanthomonas pathogens
Nature Microbiology (2024)