Abstract
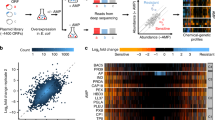
Antimicrobial peptides are promising alternative antimicrobial agents. However, little is known about whether resistance to small-molecule antibiotics leads to cross-resistance (decreased sensitivity) or collateral sensitivity (increased sensitivity) to antimicrobial peptides. We systematically addressed this question by studying the susceptibilities of a comprehensive set of 60 antibiotic-resistant Escherichia coli strains towards 24 antimicrobial peptides. Strikingly, antibiotic-resistant bacteria show a high frequency of collateral sensitivity to antimicrobial peptides, whereas cross-resistance is relatively rare. We identify clinically relevant multidrug-resistance mutations that increase bacterial sensitivity to antimicrobial peptides. Collateral sensitivity in multidrug-resistant bacteria arises partly through regulatory changes shaping the lipopolysaccharide composition of the bacterial outer membrane. These advances allow the identification of antimicrobial peptide–antibiotic combinations that enhance antibiotic activity against multidrug-resistant bacteria and slow down de novo evolution of resistance. In particular, when co-administered as an adjuvant, the antimicrobial peptide glycine-leucine-amide caused up to 30-fold decrease in the antibiotic resistance level of resistant bacteria. Our work provides guidelines for the development of efficient peptide-based therapies of antibiotic-resistant infections.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Imamovic, L. & Sommer, M. O. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci. Transl. Med. 5, 204ra132 (2013).
Lázár, V. et al. Genome-wide analysis captures the determinants of the antibiotic cross-resistance interaction network. Nat. Commun. 5, 4352 (2014).
Lázár, V. et al. Bacterial evolution of antibiotic hypersensitivity. Mol. Syst. Biol. 9, 700–700 (2013).
Toprak, E. et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat. Genet. 44, 101–105 (2012).
Palmer, A. C. & Kishony, R. Understanding, predicting and manipulating the genotypic evolution of antibiotic resistance. Nat. Rev. Genet. 14, 243–248 (2013).
Pál, C., Papp, B. & Lázár, V. Collateral sensitivity of antibiotic-resistant microbes. Trends Microbiol. 23, 401–407 (2015).
Jenssen, H., Hamill, P. & Hancock, R. E. W. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 (2006).
Fjell, C. D., Hiss, J. A., Hancock, R. E. & Schneider, G. Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51 (2012).
Mahlapuu, M., Håkansson, J., Ringstad, L. & Björn, C. Antimicrobial peptides: an emerging category of therapeutic agents. Front. Cell. Infect. Microbiol. 6, 194 (2016).
Andersson, D. I., Hughes, D. & Kubicek-Sutherland, J. Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updat. 26, 43–57 (2016).
Melo, M. N., Ferre, R. & Castanho, M. A. R. B. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 7, 245 (2009).
Alves, C. S. et al. Escherichia coli cell surface perturbation and disruption induced by antimicrobial peptides BP100 and pepR. J. Biol. Chem. 285, 27536–27544 (2010).
Avitabile, C., D’Andrea, L. D. & Romanelli, A. Circular dichroism studies on the interactions of antimicrobial peptides with bacterial cells. Sci. Rep. 4, 4293 (2014).
Soriano, F., Ponte, C., Santamaria, M. & Jimenez-Arriero, M. Relevance of the inoculum effect of antibiotics in the outcome of experimental infections caused by Escherichia coli. J. Antimicrob. Chemother. 25, 621–627 (1990).
Bechinger, B., Zasloff, M. & Opella, S. J. Structure and dynamics of the antibiotic peptide PGLa in membranes by solution and solid-state nuclear magnetic resonance spectroscopy. Biophys. J. 74, 981–987 (1998).
Ostorhazi, E., Nemes-Nikodem, E., Knappe, D. & Hoffmann, R. In vivo activity of optimized apidaecin and oncocin peptides against a multiresistant, KPC-producing Klebsiella pneumoniae strain.Prot. Pept. Lett. 21, 368–373 (2014).
Proctor, R. A. et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Micro. 4, 295–305 (2006).
Munck, C., Gumpert, H. K., Wallin, A. I., Wang, H. H. & Sommer, M. O. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci. Transl. Med. 6, 262ra156 (2014).
Mattiuzzo, M. et al. Role of the Escherichia coli SbmA in the antimicrobial activity of proline-rich peptides. Mol. Microbiol. 66, 151–163 (2007).
Pages, J.-M., James, C. E. & Winterhalter, M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Micro. 6, 893–903 (2008).
Fernández, L. & Hancock, R. E. W. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 (2012).
Seoane, A. S. & Levy, S. B. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177, 3414–3419 (1995).
Davin-Regli, A. et al. Membrane permeability and regulation of drug “influx and efflux” in enterobacterial pathogens. Curr. Drug Targets 9, 750–759 (2008).
Gutsmann, T., Fix, M., Larrick, J. W. & Wiese, A. Mechanisms of action of rabbit CAP18 on monolayers and liposomes made from endotoxins of phospholipids. J. Membr. Biol. 176, 223–236 (2000).
Kitagawa, M. et al. Complete set of ORF clones of Escherichia coli ASKA library (A complete Set of E. coli K-12 ORF Archive): unique resources for biological research. DNA Res. 12, 291–299 (2006).
Delcour, A. H. Outer membrane permeability and antibiotic resistance. BBA Proteins Proteom. 1794, 808–816 (2009).
Bociek, K. et al. Lipopolysaccharide phosphorylation by the WaaY kinase affects the susceptibility of Escherichia coli to the human antimicrobial peptide LL-37. J. Biol. Chem. 290, 19933–19941 (2015).
Lee, J.-H., Lee, K.-L., Yeo, W.-S., Park, S.-J. & Roe, J.-H. SoxRS-mediated lipopolysaccharide modification enhances resistance against multiple drugs in Escherichia coli. J. Bacteriol. 191, 4441–4450 (2009).
Gonzales, P. R. et al. Synergistic, collaterally sensitive β-lactam combinations suppress resistance in MRSA. Nat. Chem. Biol. 11, 855 (2015).
Baym, M., Stone, L. K. & Kishony, R. Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2016).
Oz, T. et al. Strength of selection pressure is an important parameter contributing to the complexity of antibiotic resistance evolution. Mol. Biol. Evol. 31, 2387–2401 (2014).
Horinouchi, T. et al. Prediction of cross-resistance and collateral sensitivity by gene expression profiles and genomic mutations. Sci. Rep. 7, 14009 (2017).
Pena-Miller, R. et al. When the most potent combination of antibiotics selects for the greatest bacterial load: The smile-frown transition. PLoS Biol. 11, e1001540 (2013).
Avan, I., Hall, C. D. & Katritzky, A. R. Peptidomimetics via modifications of amino acids and peptide bonds. Chem. Soc. Rev. 43, 3575–3594 (2014).
Jiao, Y. J., Baym, M., Veres, A. & Kishony, R. Population diversity jeopardizes the efficacy of antibiotic cycling. Preprint at https://www.biorxiv.org/content/early/2016/10/20/082107 (2016).
Fleitas, O. & Franco, O. L. Induced bacterial cross-resistance toward host antimicrobial peptides: a worrying phenomenon. Front. Microbiol. 7, 381 (2016).
Kubicek-Sutherland, J. Z. et al. Antimicrobial peptide exposure selects for Staphylococcus aureus resistance to human defence peptides. J. Antimicrob. Chemother. 72, 115–127 (2016).
Nyerges, Á. et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc. Natl Acad. Sci. 113, 2502–2507 (2016).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006 (2006).
Tenaillon, O. et al. Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536, 165 (2016).
Malouin, F., Chamberland, S., Brochu, N. & Parr, T. R. Influence of growth media on Escherichia coli cell composition and ceftazidime susceptibility. Antimicrob. Agents Chemother. 35, 477–483 (1991).
Yeaman, M. R. & Yount, N. Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 (2003).
Wiegand, I., Hilpert, K. & Hancock, R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175 (2008).
Bates, D., Machler, M., Bolker, B. M. & Walker, S. C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. https://doi.org/10.18637/jss.v067.i01 (2015).
Bolker, B. M. et al. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. 24, 127–135 (2009).
Méhi, O. et al. Perturbation of iron homeostasis promotes the evolution of antibiotic resistance. Mol. Biol. Evol. 31, 2793–2804 (2014).
Karcagi, I. et al. Indispensability of horizontally transferred genes and its impact on bacterial genome streamlining. Mol. Biol. Evol. 33, 1257–1269 (2016).
Zhou, J. & Rudd, K. E. EcoGene 3.0. Nucleic Acids Res. 41, D613–D624 (2013).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Viena, Austria, 2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Chang, D. E., Smalley, D. J. & Conway, T. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45, 289–306 (2002).
Tjaden, B. et al. Transcriptome analysis of Escherichia coli using high‐density oligonucleotide probe arrays. Nucleic Acids Res. 30, 3732–3738 (2002).
Dong, T. & Schellhorn, H. E. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol. Genet. Genom. 281, 19–33 (2009).
Notebaart, R. A. et al. Network-level architecture and the evolutionary potential of underground metabolism. Proc. Natl Acad. Sci. 111, 11762–11767 (2014).
Pierce, S. E., Davis, R. W., Nislow, C. & Giaever, G. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2, 2958–2974 (2007).
Robinson, D. G., Chen, W., Storey, J. D. & Gresham, D. Design and analysis of Bar-seq experiments. G3 4, 11–18 (2014).
Rocke, D. M. & Durbin, B. Approximate variance-stabilizing transformations for gene-expression microarray data. Bioinformatics 19, 966–972 (2003).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc. Ser. B Methodol. 57, 289–300 (1995).
Zhou, K. et al. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol. Biol. 12, 18 (2011).
Loewe, S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung 3, 285–290 (1953).
Cokol, M. et al. Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 7, 544–544 (2011).
Suzuki, S., Horinouchi, T. & Furusawa, C. Prediction of antibiotic resistance by gene expression profiles. Nat. Commun. 5, 5792 (2014).
Bódi, Z. et al. Phenotypic heterogeneity promotes adaptive evolution. PLoS Biol. 15, e2000644 (2017).
Murtagh, F. & Legendre, P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement Ward’s criterion? J. Classif. 31, 274–295 (2014).
Acknowledgements
The authors thank É. Kondorosi for providing the cationic antimicrobial peptide NCR335, and M.O.A. Sommer for the clinical isolates. The authors also acknowledge the following financial support: the Hungarian Academy of Sciences Postdoctoral Fellowship Programme (V.L.), the Hungarian Scientific Research Fund NKFI PD 116222 (A.M.), NKFI 120220 (B.K.), OTKA PD 109572 (B.C.) and NKFI FK 124254 (O.M.), the ‘Lendület’ Programme of the Hungarian Academy of Sciences and The Wellcome Trust (B.P. and C.P.), the European Research Council H2020-ERC-2014-CoG 648364 - Resistance Evolution (C.P.), GINOP-2.3.2-15-2016-00014 (EVOMER), GINOP-2.3.2-15-2016-00020 (MolMedEx TUMORDNS) and GINOP-2.3.3-15-2016-00001. I.N. and B.K. were supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences and Á.N. by the Ph.D. fellowship of the Boehringer Ingelheim Fonds.
Author contributions
C.P. and B.P. conceived and supervised the project. V.L. and A.M. designed the experiments and developed data analysis procedures. C.P., B.P., V.L., A.M. and R.S. wrote the paper. R.S., A.K., A.D., F.W. and M.D. performed the zeta potential measurements. G.O., Z.H. and T.A.M. synthetized the peptides magainin 2 and anginex. R.S. and O.M. purified RNA for transcriptomic analysis. B.B. and I.N. performed RNA-Seq experiments. P.K.J., G.F., M.S. and B.K. generated and analysed the chemogenomic data. E.U. isolated and identified E. coli clinical isolates. B.C. and A.N. prepared the mutant strains. V.L., L.D., A.M., R.S. and B.C. contributed to all other experiments. V.L., A.M., G.G., G.F. and A.G. analysed and interpreted the data.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests. B.B. and I.N. had consulting positions at SeqOmics Biotechnology Ltd. at the time the study was conceived. SeqOmics Biotechnology Ltd. was not directly involved in the design and execution of the experiments or in the writing of the manuscript. This does not alter the author’s adherence to all the Nature policies on sharing data and materials.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Text 1–4, Supplementary Methods, Supplementary Table 1, Supplementary Table 5, Supplementary Table 11, Supplementary Table 12, Supplementary Figures 1–18, Supplementary References
Supplementary Table 2
The list of antimicrobial peptides employed in this study and the available information about them based on literature mining.
Supplementary Table 3
Dataset of collateral sensitivity and cross-resistance interactions identified at the level of antibiotic-resistant strains.
Supplementary Table 4
Relative changes in the minimum inhibitory concentrations of the antimicrobial peptides towards antibiotic-resistant strains.
Supplementary Table 6
List of the main chemical and physical properties of the antimicrobial peptides employed in this study.
Supplementary Table 7
Susceptibility profiles of antibiotic-resistant E. coli clinical isolates across antimicrobial peptides.
Supplementary Table 8
Differential expression analysis of RNA-Seq data of 24 antibiotic-resistant strains.
Supplementary Table 9
Bile acid sensitivity of the antibiotic-resistant strains and list of genes involved in phospholipid and LPS synthesis.
Supplementary Table 10
List of genes sensitizing towards CAP18 and CP1 in the chemogenomic study but not to control peptide CP1.
Supplementary Table 13
Combination index (CI) values of PGLA–antibiotic (AB) combinations on E. coli clinical isolates and respective antibiotic-resistant strains.
Supplementary Table 14
Mutation-incorporating pORTMAGE oligonucleotides, allele-specific colony-PCR, HRM PCR and sequencing primers.
Rights and permissions
About this article
Cite this article
Lázár, V., Martins, A., Spohn, R. et al. Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides. Nat Microbiol 3, 718–731 (2018). https://doi.org/10.1038/s41564-018-0164-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-018-0164-0
This article is cited by
-
Antimicrobial peptide thanatin fused endolysin PA90 (Tha-PA90) for the control of Acinetobacter baumannii infection in mouse model
Journal of Biomedical Science (2024)
-
Escherichia coli resistance mechanism AcrAB-TolC efflux pump interactions with commonly used antibiotics: a molecular dynamics study
Scientific Reports (2024)
-
Synthetic peptide branched polymers for antibacterial and biomedical applications
Nature Reviews Bioengineering (2024)
-
The Synthetic Antimicrobial Peptide Derived From Melittin Displays Low Toxicity and Anti-infectious Properties
Probiotics and Antimicrobial Proteins (2024)
-
Translating eco-evolutionary biology into therapy to tackle antibiotic resistance
Nature Reviews Microbiology (2023)