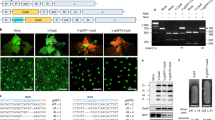
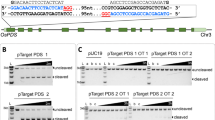
Abstract
One of the main obstacles to gene replacement in plants is efficient delivery of a donor repair template (DRT) into the nucleus for homology-directed DNA repair (HDR) of double-stranded DNA breaks. Production of RNA templates in vivo for transcript-templated HDR (TT-HDR) could overcome this problem, but primary transcripts are often processed and transported to the cytosol, rendering them unavailable for HDR. We show that coupling CRISPR-Cpf1 (CRISPR from Prevotella and Francisella 1) to a CRISPR RNA (crRNA) array flanked with ribozymes, along with a DRT flanked with either ribozymes or crRNA targets, produces primary transcripts that self-process to release the crRNAs and DRT inside the nucleus. We replaced the rice acetolactate synthase gene (ALS) with a mutated version using a DNA-free ribonucleoprotein complex that contains the recombinant Cpf1, crRNAs, and DRT transcripts. We also produced stable lines with two desired mutations in the ALS gene using TT-HDR.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
All data supporting the findings of this study are available in the article and its Supplementary Figures and Tables. Raw Sanger sequencing data are included in Supplementary Data 1.
References
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K. & Mark, A. C. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat.Commun. 7, 13274 (2016).
Puchta, H. Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J. 13, 331–339 (1998).
Cermak, T., Baltes, N. J., Cegan, R., Zhang, Y. & Voytas, D. F. High-frequency, precise modification of the tomato genome. Genome Biol. 16, 232 (2015).
Gil-Humanes, J. et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89, 1251–1262 (2017).
Sauer, N. J. et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170, 1917–1928 (2016).
Shi, J. et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216 (2017).
Sun, Y. et al. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 9, 628–631 (2016).
Wang, M. et al. Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol. Plant 10, 1007–1010 (2017).
Derr, L. K., Strathern, J. N. & Garfinkel, D. J. RNA-mediated recombination in S. cerevisiae. Cell 67, 355–364 (1991).
Nowacki, M. et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451, 153–158 (2007).
Storici, F., Bebenek, K., Kunkel, T. A., Gordenin, D. A. & Resnick, M. A. RNA-templated DNA repair. Nature 447, 338–341 (2007).
Keskin, H. et al. Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 (2014).
Chien, Y. H. & Davidson, N. RNA:DNA hybrids are more stable than DNA:DNA duplexes in concentrated perchlorate and trichloroacetate solutions. Nucleic Acids Res. 5, 1627–1637 (1978).
Butt, H. et al. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front. Plant Sci. 8, 1441 (2017).
Fonfara, I., Richter, H., Bratovič, M., Le Rhun, A. & Charpentier, E. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532, 517–521 (2016).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-cas system. Cell 163, 759–771 (2015).
Kim, D. et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 34, 863–868 (2016).
Kim, H. et al. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8, 14406 (2017).
Tang, X. et al. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 17018 (2017).
Zetsche, B. et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017).
Mazur, B. J., Chui, C. F. & Smith, J. K. Isolation and characterization of plant genes coding for acetolactate synthase, the target enzyme for two classes of herbicides. Plant Physiol. 85, 1110–1117 (1987).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Li, J., Sun, Y., Du, J., Zhao, Y. & Xia, L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 10, 526–529 (2017).
Shimatani, Z. et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 35, 441–443 (2017).
Gao, Y. & Zhao, Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J. Integr. Plant Biol. 56, 343–349 (2014).
Paix, A. et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl Acad. Sci. USA 114, E10745–E10754 (2017).
Zhang, T., Gao, Y., Wang, R. & Zhao, Y. Production of guide RNAs in vitro and in vivo for CRISPR using ribozymes and RNA polymerase II promoters. Bio Protoc. 7, e2148 (2017).
Li, L., Qu, R., de Kochko, A., Fauquet, C. & Beachy, R. N. An improved rice transformation system using the biolistic method. Plant Cell Rep. 12, 250–255 (1993).
Wang, M., Mao, Y., Lu, Y., Tao, X. & Zhu, J.-k Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 10, 1011–1013 (2017).
Hiei, Y., Ohta, S., Komari, T. & Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994).
Liu, W. et al. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol. Plant 8, 1431–1433 (2015).
Acknowledgments
We thank J.-K. Zhu for the LbCpf1 plasmid. We thank C.-Y. Wu, whose lab provided the rice transformation service. This work is partly funded by the Ministry of Agriculture and Rural Affairs of China (grant no. 2018ZX0801003B to L.X. and Y.H.), the Ministry of Science and Technology of China (grant no. 2016YFD0100500 to LX), the Ministry of Agriculture of China (grant no. 2016ZX08010003 to L.X.), and the Central Non-Profit Fundamental Research Funding supported by the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences (S2018QY05 to L.X.).
Author information
Authors and Affiliations
Contributions
L.X. and Y.Z. conceived the project. S.L., J.L., Y.H., M.X., J.Z., and W.D. performed the experiments. L.X. and Y.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have filed a patent application based on the system developed in this paper.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figures 1–5 and Supplementary Tables 1–6
Supplementary Data 1
Sanger sequence data
Rights and permissions
About this article
Cite this article
Li, S., Li, J., He, Y. et al. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol 37, 445–450 (2019). https://doi.org/10.1038/s41587-019-0065-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0065-7
This article is cited by
-
Application of multiple sgRNAs boosts efficiency of CRISPR/Cas9-mediated gene targeting in Arabidopsis
BMC Biology (2024)
-
Breeding rice for yield improvement through CRISPR/Cas9 genome editing method: current technologies and examples
Physiology and Molecular Biology of Plants (2024)
-
Amino acid substitutions in grapevine (Vitis vinifera) acetolactate synthase conferring herbicide resistance
Plant Cell, Tissue and Organ Culture (PCTOC) (2023)
-
Genome Editing Technology and Its Application to Metabolic Engineering in Rice
Rice (2022)