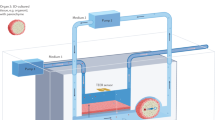
Abstract
The detection and identification of bacteria currently rely on enrichment steps such as bacterial culture and nucleic acid amplification to increase the concentration of target analytes. These steps increase assay time, cost and complexity, making it difficult to realize a truly rapid point-of-care test. Here we report the development of an electrical assay that uses electroactive RNA-cleaving DNAzymes (e-RCDs) to identify specific bacterial targets and subsequently release a DNA barcode for transducing a signal onto an electrical chip. Integrating e-RCDs into a two-channel electrical chip with nanostructured electrodes provides the analytical sensitivity and specificity needed for clinical analysis. The e-RCD assay is capable of detecting 10 CFU (equivalent to 1,000 CFU ml–1) of Escherichia coli selectively from a panel containing multiple non-specific bacterial species. Clinical evaluation of this assay using 41 patient urine samples demonstrated a diagnostic sensitivity of 100% and specificity of 78% at an analysis time of less than one hour compared with the several hours needed for currently used culture-based methods.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All relevant data presented in this study are provided in the article and its Supplementary Information. Source data are provided with this paper.
References
Klein, E. Y. et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl Acad. Sci. USA 115, E3463–E3470 (2018).
Ecker, D. J. et al. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 6, 553–558 (2008).
O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations 1–16 (Review on Antimicrobial Resistance, 2014).
Lam, B. et al. Solution-based circuits enable rapid and multiplexed pathogen detection. Nat. Commun. 4, 2001 (2013).
Davenport, M. et al. New and developing diagnostic technologies for urinary tract infections. Nat. Rev. Urol. 14, 296–310 (2017).
Nemr, C. R. Nanoparticle-mediated capture and electrochemical detection of methicillin-resistant Staphylococcus aureus. Anal. Chem. 91, 2847–2853 (2019).
Lapierre, M. A., O’Keefe, M., Taft, B. J. & Kelley, S. O. Electrocatalytic detection of pathogenic DNA sequences and antibiotic resistance markers. Anal. Chem. 75, 6327–6333 (2003).
Snodgrass, R. et al. A portable device for nucleic acid quantification powered by sunlight, a flame or electricity. Nat. Biomed. Eng. 2, 657–665 (2018).
Hung, G.-C., Nagamine, K., Li, B. & Lo, S.-C. Identification of DNA signatures suitable for use in development of real-time PCR assays by whole-genome sequence approaches: use of streptococcus pyogenes in a pilot study. J. Clin. Microbiol. 50, 2770–2773 (2012).
Young, H. et al. PCR testing of genital and urine specimens compared with culture for the diagnosis of chlamydial infection in men and women. Int. J. STD AIDS 9, 661–665 (1998).
Liu, J. et al. An integrated impedance biosensor platform for detection of pathogens in poultry products. Sci. Rep. 8, 16109 (2018).
Nidzworski, D. et al. A rapid-response ultrasensitive biosensor for influenza virus detection using antibody modified boron-doped diamond. Sci. Rep. 7, 15707 (2017).
Drummond, T. G., Hill, M. G. & Barton, J. K. Electrochemical DNA sensors. Nat. Biotechnol. 21, 1192–1199 (2003).
Heerema, S. J. & Dekker, C. Graphene nanodevices for DNA sequencing. Nat. Nanotechnol. 11, 127–136 (2016).
Li, C.-H., Chen, K.-W., Yang, C.-L., Lin, C.-H. & Hsieh, K.-C. A urine testing chip based on the complementary split-ring resonator and microfluidic channel. In 2018 IEEE Micro Electro Mechanical Systems (MEMS) 1150–1153 (IEEE, 2018); https://doi.org/10.1109/MEMSYS.2018.8346765
Point-of-Care Diagnostics Market by Testing (Glucose, Lipids, HbA1c, HCV, HIV, Influenza, Urinalysis, Hematology, Cancer, Pregnancy, PT/INR), Platform (Lateral Flow, Immunoassay), Mode (Prescription, OTC), End-User—Global Forecast to 2022 (Markets and Markets, 2018).
Li, Y. Toward an efficient DNAzyme. Biochemistry 36, 5589–5599 (1997).
Feldman, A. R., Leung, E. K. Y., Bennet, A. J. & Sen, D. The RNA-cleaving bipartite DNAzyme is a distinctive metalloenzyme. ChemBioChem 7, 98–105 (2006).
Oh, S. S., Plakos, K., Lou, X., Xiao, Y. & Soh, H. T. In vitro selection of structure-switching, self-reporting aptamers. Proc. Natl Acad. Sci. USA 107, 14053–14058 (2010).
Tang, S., Tong, P., Li, H., Tang, J. & Zhang, L. Ultrasensitive electrochemical detection of Pb2+ based on rolling circle amplification and quantum dots tagging. Biosens. Bioelectron. 42, 608–611 (2013).
Shen, L. et al. Electrochemical DNAzyme sensor for lead based on amplification of DNA–Au bio-bar codes. Anal. Chem. 80, 6323–6328 (2008).
Zhang, X.-B., Kong, R.-M. & Lu, Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 4, 105–128 (2011).
Huang, J.-Y. et al. A high-sensitivity electrochemical aptasensor of carcinoembryonic antigen based on graphene quantum dots-ionic liquid-nafion nanomatrix and DNAzyme-assisted signal amplification strategy. Biosens. Bioelectron. 99, 28–33 (2018).
Li, C., Tao, Y., Yang, Y., Xiang, Y. & Li, G. In vitro analysis of DNA–protein interactions in gene transcription using DNAzyme-based electrochemical assay. Anal. Chem. 89, 5003–5007 (2017).
Chen, J. et al. An ultrasensitive electrochemical biosensor for detection of DNA species related to oral cancer based on nuclease-assisted target recycling and amplification of DNAzyme. Chem. Commun. 47, 8004 (2011).
Xiao, Y., Rowe, A. A. & Plaxco, K. W. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J. Am. Chem. Soc. 129, 262–263 (2007).
Shamah, S. M., Healy, J. M. & Cload, S. T. Complex target SELEX. Acc. Chem. Res. 41, 130–138 (2008).
Sun, Y., Chang, Y., Zhang, Q. & Liu, M. An origami paper-based device printed with DNAzyme-containing DNA superstructures for Escherichia coli detection. Micromachines 10, 531 (2019).
Zhang, W., Feng, Q., Chang, D., Tram, K. & Li, Y. In vitro selection of RNA-cleaving DNAzymes for bacterial detection. Methods 106, 66–75 (2016).
Ali, M. M., Aguirre, S. D., Lazim, H. & Li, Y. Fluorogenic DNAzyme probes as bacterial indicators. Angew. Chem. Int. Ed. 50, 3751–3754 (2011).
Tram, K., Kanda, P., Salena, B. J., Huan, S. & Li, Y. Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem. Int. Ed. 53, 12799–12802 (2014).
Zaouri, N., Cui, Z., Peinetti, A. S., Lu, Y. & Hong, P. Y. DNAzyme-based biosensor as a rapid and accurate verification tool to complement simultaneous enzyme-based media for: E. coli detection. Environ. Sci. Water Res. Technol. 5, 2260–2268 (2019).
Flores-Mireles, A. L., Walker, J. N., Caparon, M. & Hultgren, S. J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 13, 269–284 (2015).
Zhi, L. et al. White emission magnetic nanoparticles as chemosensors for sensitive colorimetric and ratiometric detection, and degradation of ClO− and SCN− in aqueous solutions based on a logic gate approach. Nanoscale 7, 11712–11719 (2015).
Kalofonou, M. & Toumazou, C. An ISFET based analogue ratiometric method for DNA methylation detection. In 2014 IEEE International Symposium on Circuits and Systems (ISCAS) 1832–1835 (IEEE, 2014); https://doi.org/10.1109/ISCAS.2014.6865514
Yan, Y. et al. Direct ultrasensitive electrochemical biosensing of pathogenic DNA using homogeneous target-initiated transcription amplification. Sci. Rep. 6, 18810 (2016).
Rahi, A., Sattarahmady, N. & Heli, H. Zepto-molar electrochemical detection of Brucella genome based on gold nanoribbons covered by gold nanoblooms. Sci. Rep. 5, 18060 (2016).
Aguirre, S., Ali, M., Salena, B. & Li, Y. A sensitive DNA enzyme-based fluorescent assay for bacterial detection. Biomolecules 3, 563–577 (2013).
Weng, D. & Landau, U. Direct electroplating on nonconductors. J. Electrochem. Soc. 142, 2598–2604 (1995).
Mandler, D. & Bard, A. J. A new approach to the high resolution electrodeposition of metals via the feedback mode of the scanning electrochemical microscope. J. Electrochem. Soc. 137, 1079 (1990).
Werner, A. Predicting translational diffusion of evolutionary conserved RNA structures by the nucleotide number. Nucl. Acids Res. 39, e17 (2011).
Bard, A. J., Shea, T. V., Crayston, J. A., Kittlesen, G. P. & Wrighton, M. S. Digital simulation of the measured electrochemical response of reversible redox couples at microelectrode arrays: consequences arising from closely spaced ultramicroelectrodes. Anal. Chem. 58, 2321–2331 (1986).
Niwa, O., Morita, M. & Tabei, H. Electrochemical behavior of reversible redox species at interdigitated array electrodes with different geometries: consideration of redox cycling and collection efficiency. Anal. Chem. 62, 447–452 (1990).
Zhang, C. & Park, S. Simple technique for constructing thin-layer electrochemical cells. Anal. Chem. 60, 1639–1642 (1988).
Horny, M. C. et al. Electrochemical DNA biosensors based on long-range electron transfer: investigating the efficiency of a fluidic channel microelectrode compared to an ultramicroelectrode in a two-electrode setup. Lab Chip 16, 4373–4381 (2016).
Willner, I., Shlyahovsky, B., Zayats, M. & Willner, B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem. Soc. Rev. 37, 1153–1165 (2008).
Barfidokht, A. & Gooding, J. J. Approaches toward allowing electroanalytical devices to be used in biological fluids. Electroanalysis 26, 1182–1196 (2014).
Sabaté del Río, J., Henry, O. Y. F., Jolly, P. & Ingber, D. E. An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 14, 1143–1149 (2019).
Liang, J., Chen, Z., Guo, L. & Li, L. Electrochemical sensing of l-histidine based on structure-switching DNAzymes and gold nanoparticle-graphene nanosheet composites. Chem. Commun. 47, 5476–5478 (2011).
Park, K. S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 102, 179–188 (2018).
Liu, M., Chang, D. & Li, Y. Discovery and biosensing applications of diverse RNA-cleaving DNAzymes. Acc. Chem. Res. 9, 2273–2283 (2017).
Akobeng, A. K. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr. 96, 644–647 (2007).
Allen, J. B. & Larry, R. F. Electrochemical Methods: Fundamentals and Applications 2nd edn, 1505–1506 (Wiley, 2002).
Cinti, S., Moscone, D. & Arduini, F. Preparation of paper-based devices for reagentless electrochemical (bio)sensor strips. Nat. Protoc. 14, 2437–2451 (2019).
Acknowledgements
We acknowledge J. Yang for her support in SEM imaging of nanostructured gold electrodes. We thank M. Gaskin for his support towards preparing and characterizing the clinical urine samples used in this study. We acknowledge the kind help of J. Gu with the ROC plot. We thank Y. Lu for helpful discussions. The work is supported by the Natural Science and Engineering Research Council (NSERC) of Canada. L.S. and T.H. are supported by the Canada Research Chair programme. The electron microscopy was carried out at the Canadian Centre for Electron Microscopy (CCEM), a national facility supported by the NSERC and McMaster University.
Author information
Authors and Affiliations
Contributions
R.P. designed and performed electrochemical experiments and co-wrote the manuscript. D.C. designed and performed molecular experiments and edited the manuscript. M.S. supervised and helped in designing the preclinical study. T.H. supervised the assay’s performance in biological samples and edited the manuscript. Y.L. supervised the overall assay and experimental design and edited the manuscript. L.S. contributed to the overall project design and supervision and co-wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Chemistry thanks Alexis Vallée-Bélisle and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–9, Figs. 1–11, and Tables 1 and 2.
Source data
Source Data Fig. 1
Kinetics study data.
Source Data Fig. 2
Calibration plot data.
Source Data Fig. 3
Specificity data.
Source Data Fig. 4
Clinical testing data.
Source Data Fig. 5
Clinical testing data using chip-based pretreatment.
Rights and permissions
About this article
Cite this article
Pandey, R., Chang, D., Smieja, M. et al. Integrating programmable DNAzymes with electrical readout for rapid and culture-free bacterial detection using a handheld platform. Nat. Chem. 13, 895–901 (2021). https://doi.org/10.1038/s41557-021-00718-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-021-00718-x
This article is cited by
-
A wearable aptamer nanobiosensor for non-invasive female hormone monitoring
Nature Nanotechnology (2024)
-
Silver nano-reporter enables simple and ultrasensitive profiling of microRNAs on a nanoflower-like microelectrode array on glass
Journal of Nanobiotechnology (2022)
-
Host-guest liquid gating mechanism with specific recognition interface behavior for universal quantitative chemical detection
Nature Communications (2022)
-
Electrochemical aptasensor for Staphylococcus aureus by stepwise signal amplification
Microchimica Acta (2022)