Abstract
Acute lymphoblastic leukaemia (ALL) has a marked propensity to metastasize to the central nervous system (CNS). In contrast to brain metastases from solid tumours, metastases of ALL seldom involve the parenchyma but are isolated to the leptomeninges, which is an infrequent site for carcinomatous invasion. Although metastasis to the CNS occurs across all subtypes of ALL, a unifying mechanism for invasion has not yet been determined. Here we show that ALL cells in the circulation are unable to breach the blood–brain barrier in mice; instead, they migrate into the CNS along vessels that pass directly between vertebral or calvarial bone marrow and the subarachnoid space. The basement membrane of these bridging vessels is enriched in laminin, which is known to coordinate pathfinding of neuronal progenitor cells in the CNS. The laminin receptor α6 integrin is expressed in most cases of ALL. We found that α6 integrin–laminin interactions mediated the migration of ALL cells towards the cerebrospinal fluid in vitro. Mice with ALL xenografts were treated with either a PI3Kδ inhibitor, which decreased α6 integrin expression on ALL cells, or specific α6 integrin-neutralizing antibodies and showed significant reductions in ALL transit along bridging vessels, blast counts in the cerebrospinal fluid and CNS disease symptoms despite minimally decreased bone marrow disease burden. Our data suggest that α6 integrin expression, which is common in ALL, allows cells to use neural migratory pathways to invade the CNS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Garcia-Manero, G., Kantarjian, H. M. & Schiffer, C. A. in Holland-Frei Cancer Medicine (eds Kufe, D. W. et al.) (BC Decker, Hamilton, 2003).
Surapaneni, U. R. et al. Central nervous system relapse in adults with acute lymphoblastic leukemia. Cancer 94, 773–779 (2002).
Vanhaesebroeck, B., Guillermet-Guibert, J., Graupera, M. & Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell Biol. 11, 329–341 (2010).
Lannutti, B. J. et al. CAL-101, a p110δ selective phosphatidylinositol-3-kinase inhibitor for the treatment of B-cell malignancies, inhibits PI3K signaling and cellular viability. Blood 117, 591–594 (2011).
Furman, R. R. et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 370, 997–1007 (2014).
Hanna, S. & El-Sibai, M. Signaling networks of Rho GTPases in cell motility. Cell. Signal. 25, 1955–1961 (2013).
Parri, M. & Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun. Signal. 8, 23 (2010).
Reiske, H. R. et al. Requirement of phosphatidylinositol 3-kinase in focal adhesion kinase-promoted cell migration. J. Biol. Chem. 274, 12361–12366 (1999).
Webb, D. J. et al. FAK–Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6, 154–161 (2004).
Sipkins, D. A. et al. In vivo imaging of specialized bone marrow endothelial microdomains for tumour engraftment. Nature 435, 969–973 (2005).
Colmone, A. et al. Leukemic cells create bone marrow niches that disrupt the behavior of normal hematopoietic progenitor cells. Science 322, 1861–1865 (2008).
Boyerinas, B. et al. Adhesion to osteopontin in the bone marrow niche regulates lymphoblastic leukemia cell dormancy. Blood 121, 4821–4831 (2013).
Mercurio, A. M., Rabinovitz, I. & Shaw, L. M. The α6β4 integrin and epithelial cell migration. Curr. Opin. Cell Biol. 13, 541–545 (2001).
Georges-Labouesse, E., Mark, M., Messaddeq, N. & Gansmüller, A. Essential role of α6 integrins in cortical and retinal lamination. Curr. Biol. 8, 983–986 (1998).
Flanagan, L. A., Rebaza, L. M., Derzic, S., Schwartz, P. H. & Monuki, E. S. Regulation of human neural precursor cells by laminin and integrins. J. Neurosci. Res. 83, 845–856 (2006).
Belkin, A. M. & Stepp, M. A. Integrins as receptors for laminins. Microsc. Res. Tech. 51, 280–301 (2000).
DiGiuseppe, J. A., Fuller, S. G. & Borowitz, M. J. Overexpression of CD49f in precursor B-cell acute lymphoblastic leukemia: potential usefulness in minimal residual disease detection. Cytometry B Clin. Cytom. 76B, 150–155 (2009).
Coustan-Smith, E. et al. New markers for minimal residual disease detection in acute lymphoblastic leukemia. Blood 117, 6267–6276 (2011).
Terol, M.-J. et al. Expression of beta-integrin adhesion molecules in non-Hodgkin’s lymphoma: correlation with clinical and evolutive features. J. Clin. Oncol. 17, 1869–1875 (1999).
Colognato, H., ffrench-Constant, C. & Feltri, M. L. Human diseases reveal novel roles for neural laminins. Trends Neurosci. 28, 480–486 (2005).
Kienast, Y. et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat. Med. 16, 116–122 (2010).
Takei, H., Rouah, E. & Ishida, Y. Brain metastasis: clinical characteristics, pathological findings and molecular subtyping for therapeutic implications. Brain Tumor Pathol. 33, 1–12 (2016).
Wilson, E. H., Weninger, W. & Hunter, C. A. Trafficking of immune cells in the central nervous system. J. Clin. Invest. 120, 1368–1379 (2010).
Boire, A. et al. Complement component 3 adapts the cerebrospinal fluid for leptomeningeal metastasis. Cell 168, 1101–1113 (2017).
Price, R. A. & Johnson, W. W. The central nervous system in childhood leukemia. I. The arachnoid. Cancer 31, 520–533 (1973).
Leestma, J. E. Forensic Neuropathology 2nd edn (CRC, Boca Raton, 2008).
Williams, M. T. et al. The ability to cross the blood–cerebrospinal fluid barrier is a generic property of acute lymphoblastic leukemia blasts. Blood 127, 1998–2006 (2016).
Buonamici, S. et al. CCR7 signalling as an essential regulator of CNS infiltration in T-cell leukaemia. Nature 459, 1000–1004 (2009).
Kothur, K., Wienholt, L., Brilot, F. & Dale, R. C. CSF cytokines/chemokines as biomarkers in neuroinflammatory CNS disorders: a systematic review. Cytokine 77, 227–237 (2016).
Radmanesh, F. et al. Mutations in LAMB1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am. J. Hum. Genet. 92, 468–474 (2013).
Bovetti, S. et al. Blood vessels form a scaffold for neuroblast migration in the adult olfactory bulb. J. Neurosci. 27, 5976–5980 (2007).
Wade, A., McKinney, A. & Phillips, J. J. Matrix regulators in neural stem cell functions. Biochim. Biophys. Acta 1840, 2520–2525 (2014).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Acknowledgements
We thank the Duke Sequencing and Genomic Technologies (Duke Cancer Institute) and Genomic and Computational Biology shared resources and H. Dressman for gene array processing and analysis services and technical advice; M. K. Nicholas for providing expert opinions on neuro-oncology, Z. Li for assistance with statistical analysis and C. Queva for helpful discussions regarding PI3K inhibition. This work was supported by the Duke Cancer Institute and Gilead Sciences, Inc.
Reviewer information
Nature thanks I. Aifantis, J. Burger and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
D.A.S. and H.Y. designed the study and analysed the data. D.A.S. conceived and supervised the project and compiled patient data. H.Y. carried out animal experiments, pharmacokinetic studies, flow cytometry, western blotting and transwell migration assays. T.T.P. performed animal experiments, confocal microscopy and flow cytometry, analysed data and assembled figures. G.C. performed invasion assays, western blotting, flow cytometry and three-dimensional reconstructions, analysed data, assembled figures and assisted with animal experiments. B.N. and L.O. assisted with animal studies and performed histology and immunohistochemistry. M.J.W. performed microarray analyses, bone marrow intravital microscopy and histologic analyses. S.M.R. analysed data, assembled figures, performed transwell migration assays and assisted with animal experiments. E.M.J. performed intravital microscopy through thinned skull windows. S.T. and J.T. provided PI3Kδ inhibitors and assisted with pharmacokinetic analyses. C.M.M. reviewed all histology and scored immunohistochemistry staining of bone marrow biopsy samples obtained from patients. A.C. analysed data and contributed to the figures. D.A.S. wrote the manuscript. H.Y., T.T.P., G.C., S.M.R., S.T., C.M.M. and A.C. edited the paper. Key findings in the paper were replicated by a second postdoctoral researcher in the laboratory.
Corresponding author
Ethics declarations
Competing interests
D.A.S. received a grant award from Gilead Sciences, Inc. S.T. and J.T. are employees of Gilead Sciences, Inc.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 PI3Kδ inhibition minimally decreases ALL peripheral disease burden.
a, Representative haematoxylin and eosin (H&E)-stained histologic sections showing Nalm-6 disease burden in the femoral bone marrow and spleen of mice treated with vehicle or GS-649443, at matched time points. n = 6 mice per treatment group. b, Complete blood counts of vehicle- and GS-649443-treated mice, at matched time points. Paired two-tailed Student’s t-test, n = 5 mice per treatment group, P = 0.2158; ns, not significant. c, Fold changes in CNS versus systemic disease burden in vehicle- and GS-649443-treated Nalm-6-engrafted mice. Data are mean ± s.e.m.; ANOVA with Tukey’s multiple comparison test, n = 5 mice per treatment group, P = 0.0314. d, Representative histologic sections showing primary ALL disease burden in the femoral bone marrow and spleen of mice treated with vehicle or GS-649443 at matched time points. n = 5 mice per treatment group. e, Experimental plan for mice with primary ALL. f, Fold changes in CNS versus systemic disease burden in vehicle- and GS-649443-treated mice with primary ALL. Data are mean ± s.e.m.; ANOVA with Tukey, n = 5 mice per treatment group, P = 0.00128 (CNS versus bone marrow), P = 0.0098 (CNS versus spleen). g, Representative histologic sections showing RCH-ACV disease burden in the femoral bone marrow and spleen of mice treated with vehicle or GS-649443. n = 4 mice per treatment group. Arrowheads indicate RCH-ACV blasts. h, Kaplan–Meier survival curve for mice injected with RCH-ACV cells. Two-sided log rank Mantel-Cox, n = 5 mice per treatment group, P = 0.0067. i, Fold changes in CNS versus systemic disease burden in vehicle- and GS-649443-treated mice injected with RCH-ACV cells. Data are mean ± s.e.m.; ANOVA with Tukey, n = 4 mice per treatment group, P = 0.0244. Scale bars, 100 μm.
Extended Data Fig. 2 PI3Kδ inhibition at levels achievable in mouse CSF does not affect the apoptotic or cell cycle index of Nalm-6 cells; ROCK inhibition does not alter survival or disease burden in Nalm-6 leukaemic mice.
a, Serum and brain tissue concentrations of GS-649443 in healthy SCID mice. Data are mean ± s.e.m.; n = 5 mice per group. b, Effect of PI3Kδ and Akt inhibition on transwell migration of primary ALL cells. Data are mean ± s.e.m.; ANOVA with Tukey, n = 3 technical replicates per group, P = 0.5228 (vehicle versus AZD5363), P = 0.0549 (vehicle versus idelalisib), P = 0.0245 (vehicle versus GS-649443). c, Percentage of annexin-V+ Nalm-6 cells following in vitro treatment (72 h) with vehicle or GS-649443. Data are mean ± s.e.m.; unpaired two-sided Student’s t-test, n = 3 biological replicates, P = 0.2038. d, Proportion of Nalm-6 cells in the G1, S or G2/M phase of the cell cycle following in vitro treatment (72 h) with vehicle or GS-649443. Data are mean ± s.e.m.; unpaired two-sided Student’s t-test, n = 3 biological replicates, P = 0.2216 (G1), P = 0.1405 (S), P = 0.0661 (G2/M). e, Percentage of annexin-V+ cells in the CD10+ fraction of the CSF in vehicle- or GS-649443-treated mice. Paired two-sided Student’s t-test, n = 5 mice per treatment group, P = 0.6672. f, Percentage of cells in G1 in the CD10+ fraction of the CSF in vehicle- or GS-649443 treated mice. Paired two-sided Student’s t-test, n = 5 mice per treatment group, P = 0.1477. g, Percentage cells in S or G2/M phase in the CD10+ fraction of the CSF in vehicle- or GS-649443-treated mice. Paired two-sided Student’s t-test, n = 5 mice per treatment group, P = 0.5687. h, Kaplan–Meier survival curves for Nalm-6-engrafted mice treated with or without the Rho-kinase (ROCK) inhibitor fasudil. Two-sided log rank Mantel–Cox, P = 0.2843; n = 6 per treatment group. i, Disease burden (CD10+ cells) at end point in the bone marrow, spleen and CSF of vehicle- and fasudil-treated SCID mice engrafted with Nalm-6 cells. Data are mean ± s.e.m.; paired two-sided Student’s t-test, n = 4 mice per treatment group, P = 0.4534 (bone marrow), P = 0.3119 (spleen), P = 0.8026 (CSF).
Extended Data Fig. 3 PI3Kδ inhibition reduces myosin light chain activity but does not affect intra-bone marrow migration of Nalm-6 cells.
a, Western blot of MLC2 levels in ALL cells following GS-649443 treatment. For gel source data, see Supplementary Fig. 1. Nalm-6 cells: n = 3 independent experiments; primary ALL cells: n = 2 independent experiments; RCH-ACV cells: n = 3 independent experiments. b, Representative intravital microscopy images of ALL-cell intra-bone marrow migration over time in vehicle- versus GS-649443-treated mice. n = 3 mice per treatment group. Scale bars, 250 μm.
Extended Data Fig. 4 Microarray analysis of focal adhesion and motility pathway genes in blasts from vehicle- versus GS-649443-treated mice and validation of key candidates.
a, b, Differentially expressed focal adhesion and motility pathway genes identified by microarray analysis of blasts from vehicle- versus GS-649443-treated mice. n = 6 mice per treatment group. c, Microarray negative control validation. Representative flow cytometry analysis of Nalm-6 expression of CXCR4, CXCR3, CD44 and CD24 following treatment with vehicle or GS-649443. n = 3 independent experiments. d, ALL α6 integrin expression in vivo by flow cytometry. Data are mean ± s.e.m.; n = 4 mice.
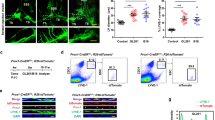
Extended Data Fig. 5 Cell surface expression of α6 integrin is variable across a panel of ALL cell lines and primary human ALL cells in vitro and in vivo.
a, Flow cytometry analysis of the percentage of α6 integrin+ populations in ALL cell lines and primary ALL cells. Data are mean ± s.e.m.; n = 3 independent experiments per cell line. b, Representative flow cytometry histograms of α6 integrin expression in ALL cell lines and primary ALL cells. n = 3 independent experiments. c, Representative α6 integrin immunohistochemistry of femoral bone marrow from mice engrafted with low α6 integrin-expressing versus high α6 integrin-expressing ALL cells. α6 integrin low primary human ALL: n = 6 mice; REH: n = 6 mice; SUP-B15: n = 3 mice; RCH-ACV: n = 6 mice; Nalm-6: n = 10 mice.
Extended Data Fig. 6 ALL cells do not invade through brain microvasculature.
a, Representative confocal microscopy images of ALL cells located within brain microvessels at various time points after intravenous engraftment. n = 3 mice per cell line and time point, 12 micrographs per mouse. White boxes outline areas shown at higher magnification in Fig. 3a. b, c, Quantification of the number of ALL cells located in brain parenchymal tissue or within brain parenchymal microvessels at various time points after intravenous engraftment of RCH-ACV (b) and REH (c) ALL cells. Data are mean ± s.e.m.; n = 3 biological replicates per time point. d, Cartoon showing vasculature within the choroid plexus, leptomeninges and brain parenchyma in humans. e, Representative GFP immunohistochemistry staining of brain parenchyma of Nalm-6–GFP-engrafted mice on day 0 after intravenous engraftment (n = 3 mice) or at end stage disease (n = 5 mice). Images show close-ups of the ALL cells that are highlighted by arrowheads in Fig. 3f. Scale bars, 100 μm.
Extended Data Fig. 7 Nalm-6 cells do not invade via diapedesis through the leptomeningeal blood–brain barrier.
a, Graphic of thinned skull window and video-rate intravital confocal microscopy approach used to image the leptomeningeal vasculature at various time points after engraftment with ALL cells. b, Representative still images from video-rate intravital microscopy analysis of leptomeningeal and superficial cerebral vasculature (red) at 10 min after intravenous Nalm-6 engraftment (full video presented in Supplementary Videos 1, 2). Nalm-6 cells (green) are observed in circulation. n = 12 mice imaged on day 0 after engraftment. c, Series of still images of leptomeningeal and superficial cerebral vasculature at 45 min after intravenous Nalm-6 engraftment. A Nalm-6 cell is observed adherent to the luminal side of a leptomeningeal vessel (white arrowhead). A second Nalm-6 cell is observed rolling along the luminal wall of a leptomeningeal vessel (blue arrowhead) (see Supplementary Video 3). No invasion via diapedesis was observed during the entirety of each 2–4-h long imaging session on the day of engraftment. n = 12 mice imaged on day 0 after engraftment. d, Still images of the leptomeningeal and superficial cerebral vasculature 12 days after Nalm-6 engraftment. No Nalm-6 cells were observed in circulation or within the leptomeningeal tissue (see Supplementary Video 4; n = 4 mice, days 2, 4, 7 and 12 after engraftment). e, Representative images from intravital confocal microscopy of the calvarial bone marrow at 2 h after Nalm-6 engraftment. Numerous Nalm-6 cells (white arrowheads) are seen to have invaded through the bone marrow vasculature soon after intravenous engraftment. n = 15 mice. f, Series of still images of the z plane of the leptomeningeal and superficial cerebral vasculature of a leukaemic mouse engrafted with Nalm-6 cells at disease end point. n = 7 mice, days 37–39 after engraftment. Nalm-6 cells are observed in circulation (white arrowheads), but no cells are observed to invade (see Supplementary Video 5).
Extended Data Fig. 8 ALL cell invasion along laminin matrices is regulated by PI3K signalling and α6 integrin.
a, Primarly ALL and Nalm-6 in vitro invasion towards human CSF along collagen, collagen and laminin, or collagen and fibronectin matrices. Data are mean ± s.e.m.; ANOVA with Tukey, n = 3 biologically independent experiments; primary ALL: P = 0.8937 (collagen versus 0.001 mg ml−1 laminin), P = 0.0020 (collagen versus 0.005 mg ml−1 laminin), P < 0.0001 (collagen versus 0.01 mg ml−1 laminin), P = 0.0604 (0.01 mg ml−4 laminin versus fibronectin); Nalm-6: P = 0.0012 (collagen versus 0.01 mg ml−1 laminin), P = 0.0150 (0.01 mg ml−1 laminin versus fibronectin). b, Comparative in vitro invasion of low α6 integrin-expressing ALL cells versus high α6 integrin-expressing ALL cells towards human CSF along laminin matrices and effects of PI3Kδ inhibition or α6 integrin blockade. Data are mean ± s.e.m.; ANOVA with Tukey, n = 3 biologically independent experiments; REH: P > 0.9999 (collagen versus laminin), P = 0.6967 (collagen and laminin versus collagen and laminin and GS-649443), P > 0.9999 (collagen and laminin versus collagen and laminin and anti-integrin-α6); primary ALL: P = 0.9758 (collagen versus laminin), P = 0.9974 (collagen and laminin versus collagen and laminin and GS-649443), P = 0.9993 (collagen and laminin versus collagen and laminin and anti-integrin-α6); RCH: P = 0.2446 (collagen versus laminin), P = 0.0079 (collagen and laminin versus collagen and laminin and GS-649443), P = 0.2549 (collagen and laminin versus collagen and laminin and anti-integrin-α6); SUP-B15: P = 0.0122 (collagen versus laminin), P = 0.0080 (collagen and laminin versus collagen and laminin and GS-649443), P = 0.0112 (collagen and laminin versus collagen and laminin and anti-integrin-α6). *P < 0.05, **P < 0.01, ***P < 0.001.
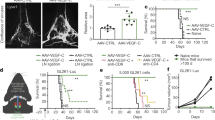
Extended Data Fig. 9 Anti-α6 integrin-blocking antibody treatment prolongs survival, but does not alter disease burden in the bone marrow or spleen of Nalm-6-engrafted leukaemic mice.
a, Schematic for the treatment of mice with the α6 integrin-neutralizing antibody. b, Kaplan–Meier survival curves for Nalm-6-engrafted mice treated with or without anti-integrin-α6 blocking antibodies. Two-sided log rank Mantel–Cox, n = 3 mice per treatment group, P = 0.0224. c, d, Disease burden at end point in the bone marrow and spleen of vehicle and anti-integrin-α6 antibody-treated Nalm-6-engrafted leukaemic mice. Data are mean ± s.e.m.; paired two-sided Student’s t-test, n = 3 mice per treatment group, P = 0.7874 (bone marrow), P = 0.1595 (spleen).
Supplementary information
Supplementary Fig. 1
This file contains Supplementary Figure 1: original source images for all data obtained by electrophoretic separation (Western Blot).
Supplementary Fig. 2
This file contains Supplementary Figure 2: gating strategy for flow cytometry data.
Video 1: Real-time in vivo imaging of leptomeninges in Nalm6-GFP leukemic mice 10 minutes post-engraftment (merged channels).
Intravital confocal microscopy performed through a thinned-skull window in a mouse approximately 10 minutes post-intravenous engraftment of Nalm6-GFP ALL cells. Circulating Nalm6-GFP cells (green) are visible in the lumen of the leptomeningeal and superficial cerebral vasculature (red), but do not extravasate. Examples of cells in circulation are best seen at time marks 00.33, 00.58, 01.03, 01.21 (mm.ss). See Supplementary Video 2 for green channel only (n = 12 mice imaged on day 0 post-engraftment).
Video 2: Real-time in vivo imaging of leptomeninges in Nalm6-GFP leukemic mice 10 minutes post-engraftment (green channel).
Intravital confocal microscopy performed through a thinned-skull window in a mouse approximately 10 minutes post-intravenous engraftment of Nalm6-GFP ALL cells. Circulating Nalm6-GFP cells (green) are visible in the lumen of the leptomeningeal and superficial vasculature (red), but do not extravasate. Examples of cells in circulation are best seen at time marks 00.33, 00.58, 01.03 and 01.21 (mm.ss). See Supplementary Video 1 for merged red and green channels (n = 12 mice imaged on day 0 post-engraftment).
Video 3: Real-time in vivo imaging of leptomeninges in Nalm6-GFP leukemic mice 45 minutes post-engraftment.
Intravital confocal microscopy performed through a thinned-skull window in a mouse approximately 45 minutes post-intravenous engraftment of Nalm6-GFP ALL cells. Nalm6-GFP cells (green) are visible attached within or circulating through the lumen of the leptomeningeal and superficial cerebral vasculature (red), but they do not extravasate. Examples of cells in circulation are best seen at time marks 00.01, 00.03 and 00.07 (mm.ss). Video play rate is slowed to facilitate viewing of cells in circulation (n = 12 mice imaged on day 0 post-engraftment).
Video 4: In vivo imaging of leptomeninges in Nalm6-GFP leukemic mice 12 days post-engraftment.
Z-stack video (10X magnification) of intravital confocal microscopy images obtained through a thinned-skull window in a mouse 12 days post-intravenous engraftment of Nalm6-GFP ALL cells. No Nalm6-GFP ALL cells (green) are detected in or around the leptomeningeal or superficial cerebral vasculature (red) (n = 4 mice imaged at intermediate time points post-engraftment).
Video 5: In vivo imaging of leptomeninges in Nalm6-GFP leukemic mice 37 days post-engraftment.
Z-stack video (20X magnification) of intravital confocal microscopy images obtained through a thinned-skull window in a mouse 37 days post-engraftment of Nalm6-GFP ALL cells. At the beginning of the Z-stack, Nalm-6-GFP+ cells (green) are visible within the BM at the edge of the skull window. In subsequent images, leptomeningeal vasculature (red) comes into focus and Nalm6-GFP ALL cells can be seen within the leptomeningeal tissue. A Nalm6-GFP cell is visible in the lumen of the CNS vasculature, but does not extravasate (timemark: 00.05 mm.ss) (n = 7 mice imaged at end stage disease).
Video 6: 3D reconstruction videos of emissary vessels within control SCID vertebral sections (example 1).
3D reconstruction video of H&E stained histologic specimens shows emissary vessels bridging the bone marrow (BM) and subarachnoid space (SA). Arrows indicate emissary vessel (n = 3 mice).
Video 7: 3D reconstruction videos of emissary vessels within control SCID vertebral sections (example 2).
3D reconstruction video of H&E stained histologic specimens shows emissary vessels bridging the bone marrow (BM) and subarachnoid space (SA). Arrows indicate emissary vessel (n = 3 mice).
Video 8: 3D reconstruction videos of Nalm-6 migration into the CNS along bone channels within vertebral sections (example 1).
3D reconstruction video of H&E stained histologic specimens from endpoint mice shows leukemic cells in transit through bony channels (arrows) bridging the BM and SA space (n = 3 mice).
Video 9: 3D reconstruction videos of Nalm-6 migration into the CNS along bone channels within vertebral sections (example 2).
3D reconstruction video of H&E stained histologic specimens from endpoint mice shows leukemic cells in transit through bony channels (arrows) bridging the BM and SA space (n = 3 mice).
Video 10: 3D reconstruction videos of RCH-ACV migration into the CNS along bone channels within vertebral sections (example 1).
3D reconstruction video of H&E stained histologic specimens from endpoint mice shows leukemic cells in transit through bony channels (arrows) bridging the BM and SA space (n = 3 mice).
Video 11: 3D reconstruction videos of RCH-ACV migration into the CNS along bone channels within vertebral sections (example 2).
3D reconstruction video of H&E stained histologic specimens from endpoint mice shows leukemic cells in transit through bony channels (arrows) bridging the BM and SA space (n = 3 mice).
Source data
Rights and permissions
About this article
Cite this article
Yao, H., Price, T.T., Cantelli, G. et al. Leukaemia hijacks a neural mechanism to invade the central nervous system. Nature 560, 55–60 (2018). https://doi.org/10.1038/s41586-018-0342-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0342-5
This article is cited by
-
Role of meningeal immunity in brain function and protection against pathogens
Journal of Inflammation (2024)
-
Decoding leukemia at the single-cell level: clonal architecture, classification, microenvironment, and drug resistance
Experimental Hematology & Oncology (2024)
-
Targeting cytohesin-1 suppresses acute myeloid leukemia progression and overcomes resistance to ABT-199
Acta Pharmacologica Sinica (2024)
-
Imaging of brain barrier inflammation and brain fluid drainage in human neurological diseases
Cellular and Molecular Life Sciences (2024)
-
Identification of direct connections between the dura and the brain
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.