Abstract
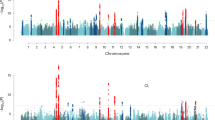
The genetic architecture of human reproductive behavior—age at first birth (AFB) and number of children ever born (NEB)—has a strong relationship with fitness, human development, infertility and risk of neuropsychiatric disorders. However, very few genetic loci have been identified, and the underlying mechanisms of AFB and NEB are poorly understood. We report a large genome-wide association study of both sexes including 251,151 individuals for AFB and 343,072 individuals for NEB. We identified 12 independent loci that are significantly associated with AFB and/or NEB in a SNP-based genome-wide association study and 4 additional loci associated in a gene-based effort. These loci harbor genes that are likely to have a role, either directly or by affecting non-local gene expression, in human reproduction and infertility, thereby increasing understanding of these complex traits.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Elks, C.E. et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 42, 1077–1085 (2010).
Perry, J.R.B. et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature 514, 92–97 (2014).
Rahmioglu, N. et al. Genetic variants underlying risk of endometriosis: insights from meta-analysis of eight genome-wide association and replication datasets. Hum. Reprod. Update 20, 702–716 (2014).
Day, F.R. et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat. Commun. 6, 8464 (2015).
Mehta, D. et al. Evidence for genetic overlap between schizophrenia and age at first birth in women. JAMA Psychiatry 73, 497–505 (2016).
Mills, M.C. & Tropf, F.C. The biodemography of fertility: a review and future research frontiers. Kolner Z. Soz. Sozpsychol. 67 (Suppl. 1), 397–424 (2015).
Mills, M., Rindfuss, R.R., McDonald, P. & te Velde, E. Why do people postpone parenthood? Reasons and social policy incentives. Hum. Reprod. Update 17, 848–860 (2011).
Boivin, J., Bunting, L., Collins, J.A. & Nygren, K.G. International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical care. Hum. Reprod. 22, 1506–1512 (2007).
Mascarenhas, M.N., Flaxman, S.R., Boerma, T., Vanderpoel, S. & Stevens, G.A. National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med. 9, e1001356 (2012).
Venkatesh, T., Suresh, P.S. & Tsutsumi, R. New insights into the genetic basis of infertility. Appl. Clin. Genet. 7, 235–243 (2014).
Day, F.R. et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat. Genet. 48, 617–623 (2016).
Balbo, N., Billari, F.C. & Mills, M.C. Fertility in advanced societies: a review of research. Eur. J. Popul. 29, 1–38 (2012).
Tropf, F.C. et al. Human fertility, molecular genetics, and natural selection in modern societies. PLoS One 10, e0126821 (2015).
Fisher, R.A. The Genetical Theory of Natural Selection (Oxford University Press, 1930).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
van der Most, P.J. et al. QCGWAS: a flexible R package for automated quality control of genome-wide association results. Bioinformatics 30, 1185–1186 (2014).
Winkler, T.W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Lango Allen, H. et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467, 832–838 (2010).
Bulik-Sullivan, B.K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Purcell, S.M. et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009).
Liu, J.Z. et al. A versatile gene-based test for genome-wide association studies. Am. J. Hum. Genet. 87, 139–145 (2010).
Mishra, A. & Macgregor, S. VEGAS2: software for more flexible gene-based testing. Twin Res. Hum. Genet. 18, 86–91 (2015).
Vaez, A. et al. In silico post genome-wide association studies analysis of C-reactive protein loci suggests an important role for interferons. Circ Cardiovasc Genet 8, 487–497 (2015).
ENCODE Project Consortium. ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 (2004).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Boyle, A.P. et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Zhernakova, D. et al. Hypothesis-free identification of modulators of genetic risk factors. Preprint at bioRxiv http://dx.doi.org/10.1101/033217 (2015).
Bonder, M.J. et al. Disease variants alter transcription factor levels and methylation of their binding sites. Preprint at bioRxiv http://dx.doi.org/10.1101/033084 (2015).
Tranchevent, L.C. et al. ENDEAVOUR update: a web resource for gene prioritization in multiple species. Nucleic Acids Res. 36, W377–W384 (2008).
Pers, T.H., Dworzyn´ski, P., Thomas, C.E., Lage, K. & Brunak, S. MetaRanker 2.0: a web server for prioritization of genetic variation data. Nucleic Acids Res. 41, W104–W108 (2013).
Chen, J., Bardes, E.E., Aronow, B.J. & Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 37, W305–W311 (2009).
Pers, T.H. et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat. Commun. 6, 5890 (2015).
Euesden, J., Lewis, C.M. & O'Reilly, P.F. PRSice: Polygenic Risk Score software. Bioinformatics 31, 1466–1468 (2015).
Okbay, A. et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016).
Willer, C.J. et al. Discovery and refinement of loci associated with lipid levels. Nat. Genet. 45, 1274–1283 (2013).
Locke, A.E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Rietveld, C.A. et al. Common genetic variants associated with cognitive performance identified using the proxy-phenotype method. Proc. Natl. Acad. Sci. USA 111, 13790–13794 (2014).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Day, F.R. et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat. Genet. 47, 1294–1303 (2015).
Perry, J.R. et al. A genome-wide association study of early menopause and the combined impact of identified variants. Hum. Mol. Genet. 22, 1465–1472 (2013).
Fang, W.-L. et al. CREB coactivator CRTC2/TORC2 and its regulator calcineurin crucially mediate follicle-stimulating hormone and transforming growth factor β1 upregulation of steroidogenesis. J. Cell. Physiol. 227, 2430–2440 (2012).
Cao, G. et al. Molecular cloning and characterization of a novel human cAMP response element–binding (CREB) gene (CREB4). J. Hum. Genet. 47, 373–376 (2002).
El-Alfy, M. et al. Stage-specific expression of the Atce1/Tisp40α isoform of CREB3L4 in mouse spermatids. J. Androl. 27, 686–694 (2006).
Adham, I.M. et al. Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol. Cell. Biol. 25, 7657–7664 (2005).
McAllister, J.M. et al. Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc. Natl. Acad. Sci. USA 111, E1519–E1527 (2014).
O'Bryan, M.K. et al. RBM5 is a male germ cell splicing factor and is required for spermatid differentiation and male fertility. PLoS Genet. 9, e1003628 (2013).
Tsukamoto, S. et al. Functional analysis of lysosomes during mouse preimplantation embryo development. J. Reprod. Dev. 59, 33–39 (2013).
Szucs, M., Osvath, P., Laczko, I. & Jakab, A. Adequacy of hyaluronan binding assay and a new fertility index derived from it for measuring of male fertility potential and the efficacy of supplement therapy. Andrologia 47, 519–524 (2015).
Buensuceso, A.V. et al. Ephrin-A5 is required for optimal fertility and a complete ovulatory response to gonadotropins in the female mouse. Endocrinology 157, 942–955 (2016).
Jisa, E. & Jungbauer, A. Kinetic analysis of estrogen receptor homo- and heterodimerization in vitro. J. Steroid Biochem. Mol. Biol. 84, 141–148 (2003).
O'Donnell, L., Robertson, K.M., Jones, M.E. & Simpson, E.R. Estrogen and spermatogenesis. Endocr. Rev. 22, 289–318 (2001).
Ly-Huynh, J.D. et al. Importin α2–interacting proteins with nuclear roles during mammalian spermatogenesis. Biol. Reprod. 85, 1191–1202 (2011).
Varshney, G.K. et al. CRISPRz: a database of zebrafish validated sgRNAs. Nucleic Acids Res. 44, D1, D822–D826 (2016).
Menken, J. Age and fertility: how late can you wait? Demography 22, 469–483 (1985).
Manolio, T.A., Brooks, L.D. & Collins, F.S. A HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008).
Hindorff, L.A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA 106, 9362–9367 (2009).
Okkelman, I.A., Sukaeva, A.Z., Kirukhina, E.V., Korneenko, T.V. & Pestov, N.B. Nuclear translocation of lysyl oxidase is promoted by interaction with transcription repressor p66β. Cell Tissue Res. 358, 481–489 (2014).
Joshi, N.R. et al. Altered expression of microRNA-451 in eutopic endometrium of baboons (Papio anubis) with endometriosis. Hum. Reprod. 30, 2881–2891 (2015).
Franklin, R.B. et al. Human ZIP1 is a major zinc uptake transporter for the accumulation of zinc in prostate cells. J. Inorg. Biochem. 96, 435–442 (2003).
Lisle, R.S., Anthony, K., Randall, M.A. & Diaz, F.J. Oocyte–cumulus cell interactions regulate free intracellular zinc in mouse oocytes. Reproduction 145, 381–390 (2013).
Shan, B. et al. Association of DENND1A gene polymorphisms with polycystic ovary syndrome: a meta-analysis. J. Clin. Res. Pediatr. Endocrinol. 8, 135–143 (2016).
Impera, L. et al. A novel fusion 5′AFF3/3′BCL2 originated from a t(2;18)(q11.2;q21.33) translocation in follicular lymphoma. Oncogene 27, 6187–6190 (2008).
Urano, A. et al. Infertility with defective spermiogenesis in mice lacking AF5q31, the target of chromosomal translocation in human infant leukemia. Mol. Cell. Biol. 25, 6834–6845 (2005).
Reese, K.L. et al. Acidic hyaluronidase activity is present in mouse sperm and is reduced in the absence of SPAM1: evidence for a role for hyaluronidase 3 in mouse and human sperm. Mol. Reprod. Dev. 77, 759–772 (2010).
Heath, E., Sablitzky, F. & Morgan, G.T. Subnuclear targeting of the RNA-binding motif protein RBM6 to splicing speckles and nascent transcripts. Chromosome Res. 18, 851–872 (2010).
Kamura, T. et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6, 1229–1235 (2004).
Kato, J.Y., Matsuoka, M., Polyak, K., Massagué, J. & Sherr, C.J. Cyclic AMP–induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell 79, 487–496 (1994).
Bagley, D.C., Paradkar, P.N., Kaplan, J. & Ward, D.M. Mon1a protein acts in trafficking through the secretory apparatus. J. Biol. Chem. 287, 25577–25588 (2012).
Sakamoto, O. et al. Role of macrophage-stimulating protein and its receptor, RON tyrosine kinase, in ciliary motility. J. Clin. Invest. 99, 701–709 (1997).
Zhang, C. et al. Molecular mechanisms that drive estradiol-dependent burst firing of Kiss1 neurons in the rostral periventricular preoptic area. Am. J. Physiol. Endocrinol. Metab. 305, E1384–E1397 (2013).
Ponglikitmongkol, M., Green, S. & Chambon, P. Genomic organization of the human oestrogen receptor gene. EMBO J. 7, 3385–3388 (1988).
de Mattos, C.S. et al. ESR1 and ESR2 gene polymorphisms are associated with human reproduction outcomes in Brazilian women. J. Ovarian Res. 7, 114 (2014).
Lamp, M. et al. Polymorphisms in ESR1, ESR2 and HSD17B1 genes are associated with fertility status in endometriosis. Gynecol. Endocrinol. 27, 425–433 (2011).
Chiu, Y.-C. et al. Foxp2 regulates neuronal differentiation and neuronal subtype specification. Dev. Neurobiol. 74, 723–738 (2014).
Alves, M.G. et al. Metabolic fingerprints in testicular biopsies from type 1 diabetic patients. Cell Tissue Res. 362, 431–440 (2015).
Mojiminiyi, O.A., Safar, F.H., Al Rumaih, H. & Diejomaoh, M. Variations in alanine aminotransferase levels within the normal range predict metabolic and androgenic phenotypes in women of reproductive age. Scand. J. Clin. Lab. Invest. 70, 554–560 (2010).
Van Maldergem, L. et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller–Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 43, 148–152 (2016).
Ruan, Y., Cheng, M., Ou, Y., Oko, R. & van der Hoorn, F.A. Ornithine decarboxylase antizyme Oaz3 modulates protein phosphatase activity. J. Biol. Chem. 286, 29417–29427 (2011).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Zhu, Z. et al. Dominance genetic variation contributes little to the missing heritability for human complex traits. Am. J. Hum. Genet. 96, 377–385 (2015).
Shen, X. et al. Simple multi-trait analysis identifies novel loci associated with growth and obesity measures. Preprint at bioRxiv http://dx.doi.org/10.1101/022269 (2015).
Okbay, A. et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016).
Winkler, T.W. et al. EasyStrata: evaluation and visualization of stratified genome-wide association meta-analysis data. Bioinformatics 31, 259–261 (2015).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C. & Morris, Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9 (Suppl. 1), S4 (2008).
Saito, R. et al. A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 (2012).
Montojo, J. et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 (2010).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Kohler, H.-P., Rodgers, J.L. & Christensen, K. Is fertility behavior in our genes? Findings from a Danish twin study. Popul. Dev. Rev. 25, 253–288 (1999).
Tropf, F.C., Barban, N., Mills, M.C., Snieder, H. & Mandemakers, J.J. Genetic influence on age at first birth of female twins born in the UK, 1919–68. Popul. Stud. (Camb.) 69, 129–145 (2015).
Voight, B.F., Kudaravalli, S., Wen, X. & Pritchard, J.K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Acknowledgements
For full acknowledgments, see the Supplementary Note. Funding to lead this consortium was provided by grants awarded to M.C.M.: ERC Consolidator Grant SOCIOGENOME (615603), a Dutch Science Foundation (NWO) grant (VIDI grant 452-10-012), a UK ESRC/NCRM SOCGEN grant (ES/N011856/1), the European Union's FP7 Families And Societies project (320116), and the Wellcome Trust ISSF and John Fell Fund. M.d.H. was supported by grants from the Swedish Research Council (2015-03657) and the Swedish Heart-Lung Foundation (20140543). Research was carried out in collaboration with the Social Science Genetic Association Consortium (SSGAC), with funding from the US National Science Foundation (EAGER: 'Workshop for the Formation of a Social Science Genetic Association Consortium'), a Supplementary grant from the National Institutes of Health Office of Behavioral and Social Science Research, the Ragnar Söderberg Foundation (E9/11), the Swedish Research Council (421-2013-1061), the Jan Wallander and Tom Hedelius Foundation, an ERC Consolidator Grant (647648 EdGe), the Pershing Square Fund of the Foundations of Human Behavior and the NIA/NIH through grants P01-AG005842, P01-AG005842-20S2, P30-AG012810 and T32-AG000186-23 to NBER and R01-AG042568-02 to the University of Southern California. X.S. was supported by a grant from the Swedish Research Council (537-2014-371). We thank X. Ding for research assistance, N. Pirastu, K. Coward and L. Layman for valuable comments, and the University of Oxford Advanced Research Computing (ARC) facility (http://dx.doi.org/10.5281/zenodo.22558). This research has been conducted using the UK Biobank Resource.
Author information
Authors and Affiliations
Consortia
Contributions
Senior investigators who led writing, analysis and study design: M.C.M., H. Snieder and M.d.H. Senior investigators who participated in study design: P.D.K., D.J.B. and D.C. Junior investigator who contributed to the study design and management: N. Barban. Population stratification: N. Barban and F.C.T. Genetic correlations and polygenic score prediction: N. Barban. Meta-analysis and quality control: N. Barban, R.d.V., J.J.M. and I.M.N. Biological annotation: R.J., M.d.H. and A.V. Sex-specific genetic effects: N. Barban and F.C.T. Bivariate and conditional analysis of the two fertility traits: X.S., J.F.W. and D.I.C. Gene-based analysis V.T. and S.W.v.d.L. Authors not listed contributed to recruitment, genotyping or data processing for the meta-analysis (Supplementary Table 43).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–37 and Supplementary Note. (PDF 11303 kb)
Supplementary Tables 1–43
Supplementary Tables 1–43. (XLS 12508 kb)
Rights and permissions
About this article
Cite this article
Barban, N., Jansen, R., de Vlaming, R. et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat Genet 48, 1462–1472 (2016). https://doi.org/10.1038/ng.3698
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3698
This article is cited by
-
The impact of reproductive factors on the metabolic profile of females from menarche to menopause
Nature Communications (2024)
-
Risk factors for human papillomavirus infection, cervical intraepithelial neoplasia and cervical cancer: an umbrella review and follow-up Mendelian randomisation studies
BMC Medicine (2023)
-
Establishing the relationships between adiposity and reproductive factors: a multivariable Mendelian randomization analysis
BMC Medicine (2023)
-
Genome-wide analysis identifies genetic effects on reproductive success and ongoing natural selection at the FADS locus
Nature Human Behaviour (2023)
-
The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions
Nature Genetics (2023)