Abstract
Triterpenic acids (TAs), a large group of natural compounds with diverse biological activity, are produced by several plant taxa. Betulinic, oleanolic, and ursolic acids are the most medicinally important TAs and are mainly found in plants of the mint family. Metabolic engineering is strongly dependent on identifying the key genes in biosynthetic pathways toward the products of interest. In this study, gene expression tracking was performed by transcriptome mining, co-expression network analysis, and tissue-specific metabolite-expression analysis in order to identify possible key genes involved in TAs biosynthetic pathways. To this end, taxa-specific degenerate primers of six important genes were designed using an effective method based on the MEME algorithm in a phylogenetically related group of sequences and successfully applied in three members of the Lamiaceae (Rosmarinus officinalis, Salvia officinalis, and Thymus persicus). Based on the results of in-depth data analysis, genes encoding squalene epoxidase and oxido squalene cyclases are proposed as targets for boosting triterpene production. The results emphasize the importance of identifying key genes in triterpene biosynthesis, which may facilitate genetic manipulation or overexpression of target genes.
Similar content being viewed by others
Introduction
Terpenoids are a group of natural compounds derived from five-carbon isoprene units and are classified according to the number and structural organization of these biosynthetic building blocks. Among the different structural categories (mono-, sesqui-, di-, tri, tetra, and polyterpenoids), the largest and most diverse group are the triterpenoids (TTs, C30H48), with a wide distribution in plants1. These organic compounds can be further subdivided into subclasses based on the number of available rings in the structure. Three well-known triterpenic acids (TAs) of the lupane, oleanane and ursane type, namely betulinic acid (3β-hydroxylup-20(29)-en-28-oic acid, BA), oleanolic acid (3β-hydroxyolean-12-en-28-oic acid, OA), and ursolic acid (3β-hydroxyurs-12-en-28-oic acid, UA), have been frequently reported from different plant organs such as leaves, flowers, berries, and fruits2,3,4. Various biological effects of TAs including anti-inflammatory and antioxidant5, anti-HIV6, anti-fungal and immunomodulatory activities7,8 have been characterized. Additionally, extensive bioassays of TAs against different human cancer cell lines have been performed9,10,11,12,13,14,15,16. Recently, potential effects of BA, OA, and UA for the treatment of type 2 diabetes (T2DM) have been highlighted17. Therefore, due to this considerable pharmacological activity and potential high demand for these medicinally important compounds, there is an urgent need to find the best natural source and biotechnological approach for their production. TAs have been reported from a range of plant families including Dilleniaceae18, Ericaceae19,20, Euphorbiaceae21, Rosaceae22 and Myrtaceae23. The mint family (Lamiaceae) is considered to be a particularly rich source of TAs24, which have been isolated and quantitatively determined in Salvia species25,26,27, Rosmarinus officinalis26, Thymus species28, Prunella vulgaris29, Orthosiphon stamineus30, and Melissa officinalis24, among others.
Plant isoprenoids are derived from the cytoplasmic mevalonate (MVA) and the plastidial methylerythritol phosphate (MEP) pathways, which generate the isoprenoid precursors dimethylallyl pyrophosphate (DMAPP) and isopentenyl pyrophosphate (IPP). TAs are biosynthetically produced through the cyclization of squalene31, a hydrocarbon reported to be the precursor of all isoprenoids in eukaryotes. After the conversion of squalene to 2,3-oxidosqualene, cyclization by specific oxidosqualene cyclases (OSCs) generates a wide variety of triterpenoid backbones, including lupeol, β-amyrin, and α-amyrin, which are decorated by multiple cytochrome P450-dependent monooxygenases (P450s) to produce BA, OA and UA, respectively32,33. The main steps of TAs biosynthesis are shown in Fig. 1.
The biosynthetic route to pentacyclic triterpenes of betulinic, oleanolic and ursolic acids. These compounds are synthesized via the mevalonic acid (MVA) and methylerythritol 4-phosphate (MEP) pathway. The MVA pathway enzymes are shown on the left and the MEP pathway enzymes on the right in a blue box. The enzymes that catalyze the various steps are indicated in ovals. Enzyme abbreviations: DXS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CMK, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol kinase; MDS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase; HMGS, 3-hydroxy-3-methylglutaryl coenzyme A synthase; HMGR, 3-hydroxy-3-methylglutaryl coenzyme a reductase; MK, Mevalonate kinase; PMK, phosphomevalonate kinase; MDC, mevalonate-5-decarboxylase; IDI, isopentenyl-diphosphate isomerase; GPPS, geranyl diphosphate synthase; FDS, farnesyl pyrophosphate synthase; SQS, squalene synthase; SQE, squalene monooxygenase or epoxidase; OSCs, oxido squalene cyclases.
A large number of studies have shown a strong link between enzyme expression and metabolite production34,35,36,37, although the role of other regulatory mechanisms, including transcriptional, translational and post-translational regulation, is still unclear. In addition, the whole transcriptome should be considered, as the final product of a metabolic pathway is determined by a series of gene actions. High-throughput transcriptomics produces extensive transcript datasets that can be applied to identify candidate key genes in particular processes using co-expression networks analysis38,39. Examples include the identification of genes involved in disease40, cancer41,42,43, pathogen resistance39,44,45, different organs or developmental stages46,47,48 and biotic and abiotic stress in plants49,50,51. After the crucial genes are identified, the sequences encoding TAs biosynthetic enzymes need to be elucidated for molecular applications such as metabolic engineering. In the candidate gene approach, genes exhibiting a significant sequence similarity, involved in the same biological process so they can amplify by well-designed degenerate primers in the related organisms52. Targeted sequencing by these primers reduces the cost and time of sequencing projects and enables a more in-depth evaluation of key biosynthetic genes. Successful amplification requires the design of universal degenerate primers for members of a particular plant family, an example being the amplification of a key gene encoding enzymes involved in proanthocyanidins (PAs) biosynthesis in the Rosaceae family53.
There is a lack of genomic data for non-model plants, especially medicinal plants, but in silico expression analysis can bridge the knowledge gap and facilitate the identification of the main genes affecting the production of secondary metabolites (SMs). In the present study, the expression profiles of mRNA related to different organs in plants in the mint family under elicitation were analyzed for in silico studies. Co-expression network analysis was used to identify the key genes and their expression pattern in the TAs biosynthetic pathway. The genes were then amplified using designed degenerate primers in selected members of the mint family. Finally, biosynthetic genes related to BA, OA and UA production were analyzed by real-time polymerase chain reaction (RT-PCR), and high-performance liquid chromatography (HPLC) was performed to check whether the transcript levels were directly related to the TAs content. The results provide valuable information for biotechnological applications aimed at improving TAs production in plants of the mint family.
Results
Highlighting the expression pattern of TAs biosynthetic genes from high-throughput expression data
Transcriptome analysis experiments provide researchers with a snapshot of a subset of the expressed target genes under various conditions. The current study focused on the expression pattern of TAs biosynthetic genes through analyzing the gene expression profiles of Lamiaceae plants obtained from high-throughput transcriptome profiling experiments. In order to gain more insight into the TAs biosynthetic pathway, expression data sets were analyzed, and differentially expressed genes (DEGs) were identified using the BLAST algorithm, and a co-expression network was constructed to distinguish the key genes in the pathway. The retrieved expression data of TAs genes were analyzed, clustered based on the similarity of their expression pattern, and visualized by R software.
As shown in Fig. 2, a relationship was observed between the gene expression values and the specific tissue/elicitor. In general, biosynthetic genes in plants show a spatio-temporal expression pattern (particular tissue /developmental stage). In addition, these genes can be induced by stressful conditions or by elicitors such as MeJA54,55,56. Hierarchical clustering of gene expression values revealed two main clusters (Fig. 2b,c). If the genes involved in TAs biosynthesis are divided into upstream (MVA and MEP pathway) and downstream (FDS, SQS, SQE, OSCs), a downstream biosynthetic gene cluster is apparent, and most of the genes are overexpressed under elicitation compared with upstream genes (Fig. 2c). The co-expression network of upstream and downstream TAs biosynthetic genes was analyzed with network parameters, as illustrated in Fig. 3. Nodes are considered as the genes, and edges indicate the magnitude of their co-expression. A scatter-plot of the betweenness centrality (BC) versus degree (Fig. 3b) was generated to identify genes with high relative values in both parameters. MVD1 (mevalonate diphosphate decarboxylase, MDC in Fig. 1), AT3G54250 (GHMP kinase family proteins), OSCs and SQE showed high values based on these two parameters. Further, these nodes ranked highly according to other centrality parameters of closeness, stress, and radiality. However, when considering eigenvector parameters, the OSCs and SQE transformed the top ranking. The eigenvector centrality measures the influence of a node on the network. Arabidopsis MVD is encoded by two loci, and is responsible for the last step of the MVA pathway where mevalonic acid is converted into isopentenyl diphosphate (IPP). Regarding the co-expression relationship of downstream genes with the TAs pathway, SQE co-expresses with different triterpene cyclases (OSCs). Triterpene cyclases use 2,3-oxidosqualene as a substrate, a common biosynthetic intermediate for plant triterpenes, and convert it into a variety of pentacyclic triterpene scaffolds. OSCs can be multifunctional or produce only one product. Lupane, oleanane, and ursane skeletons are catalyzed by lupeol synthase (LUS), β-amyrin synthase (BAS), and α-amyrin (AAS) synthase or mixed-function amyrin synthase (MFAS) as OCSs, respectively. The pentacyclic skeletons are usually modified further to produce BA, OA and UA by cytochromes P450, acetyltransferases and glycosyltransferases57.
Heat map showing the expression profiles of TAs genes in four series of high-throughput sequencing data. (a) SRP006813- RNA sequencing of Rosmarinus officinalis (different tissues), (b) SRP126897- RNA sequencing of Salvia splendens (different tissues), (c) SRP111399- Transcriptome profiling of Salvia miltiorrhiza (elicited by methyl jasmonate (MeJA) and yeast extract (YE)) (d) SRP065934- Transcriptome profiling of Salvia miltiorrhiza (elicited by salicylic acid (SA)). Transcripts for some genes involved in TAs biosynthesis were not detected in experiment 1 because of the low size of reads, and so these genes were not included.
Bioinformatics analysis to design degenerate primers and RT-PCR
RNA-Seq provides a unique combination of transcriptome which can be considered as a repository for selecting the targets of interest. To design degenerate specific taxa primers for validating the expression patterns of TAs genes, nucleotide sequences of FDS, SQS, SQE, and OSCs (LUP, BAS, and MFAS) were retrieved from the transcriptome of rosemary (SRP006813). The predicted amino acid sequences were then subjected to phylogenetic analysis and the results (Fig. 4) indicate that all proteins of the TAs pathway can be divided into four large groups (I, II, III, and IV). Among these groups, branch IV consisted of OSCs and was further divided into three subgroups (LUS, BAS, and MFAS). These enzymes exhibited a much closer evolutionary relationship with each other. The reaction pathways by which TAs are generated via oxidosqualene cyclization catalyzed by the OSC family are divergent in nature. Members of this enzyme family have a very high sequence identity, which poses a challenge for determining exactly how each OSC controls the product specificity. However, some OSCs are multifunctional and normally convert 2,3-oxidosqualene into a variety of products. For example, MFAS cyclizes 2,3-oxidosqualene to α-amyrin and β-amyrin in a 5:3 ratio, which was demonstrated in experiments in yeast in which overexpression of MFAS in Ocimum basilicum led to the preferential production of α-amyrin56. Based on the sequence analysis, BAS and MFAS in this plant shared 73% identity, indicating that only a slight difference in their sequences may cause differences in the catalytic functions of OSCs.
Phylogenetic analysis of TAs biosynthesis enzymes. The amino acid sequences were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). Predicted proteins of R. officinalis are shown in red. The tree was constructed using MEGA 7.0 software with an evaluation method of Bootstrap repeated a 1000 times. The scale bar represents the number of amino acid substitutions per site.
Predicted proteins of LUS, BAS, and MFAS of R. officinalis were more phylogenetically related to LUS from Taraxacum officinalis and Artemisia annua, and BAS and MFAS from O. basilicum, sharing 83%, 78%, 88%, and 90% identity at the amino acid sequence level, respectively. As depicted in Fig. 4, the branching pattern of the phylogenetic tree reflected that the predicted amino acid sequences of FDS, SQS, and SQE were phylogenetically related to these enzymes in Salvia, the most closely related species to R. officinalis in the mint family, with the highest sequence identity (96%), and evolving from a common ancestor. The amino acid sequences of species closely related to R. officinalis according to the phylogeny tree were aligned, and the conserved blocks of sequences were identified by the Multiple Expectation – Maximization for Motif Elicitation (MEME) algorithm. Figure 5 displays the motif pairs used to design the primer pairs for each gene. Table 1 indicates the designed degenerate primers that passed the specificity checking process.
In order to illustrate the performance of the designed primer in amplifying target fragments of the TAs biosynthetic genes from Lamiaceae plants, a two-step RT-PCR was performed and gel electrophoresis was used to visualize the results (Fig. 6). Based on the results, a single DNA product matched the expected size of the PCR amplicon for each gene in three members of the Lamiaceae family, which indicated that the primers worked efficiently amplified the amplicons produced by degenerating specific taxa primers. However, MFAS primers failed to amplify the responding gene in T. persicus and did not show any band at all, maybe because the sequence of this gene was available only in a few plants and it was not sufficient for designing universal primers.
Subsequently, the amplicons were purified, sequenced, and then analyzed by bioinformatics tools to confirm the results of the RT-PCR analysis. In addition, all of the sequences were successfully affiliated and corresponded to the target genes of the TAs biosynthetic pathway according to the BLAST sequence analysis.
Tissue-specific gene expression and TAs Profile
In order to determine the tissue-dependent expression of TAs enzymes, a panel of 6 genes, FDS, SQS, SQE, LUS, BAS, and MFAS, were profiled in four tissues: leaf, flower, stem, and root. The average contents of BA, OA, and UA in different tissues of R. officinalis, S. officinalis, and T. persicus were determined using the standard equations and expressed in mg/g dry weight (DW) of the material (Fig. 7a). Although a large body of phytochemical research on TAs evaluation has been quantitatively conducted in plants, including different species of the Lamiaceae family26,27, no studies have focused on the TAs profile in different tissues of these plants except Salvia58. Tissue-specific gene expression and TAs profile is reported in the present study for the first time.
Tissue-specific changes in TAs biosynthesis in metabolite and transcript levels. (a) Estimation of TAs contents (mg/g DW) in different tissues of three members of the Lamiaceae family. The bars represent total measured TAs and the panel are means ± SD. Significance levels for the difference between any pair of means (TAs content) by Duncan’s multiple range test are marked above the data in lowercase. (b) Transcript abundance of TAs biosynthetic genes, relative transcript levels of farnesyl pyrophosphate synthase (FDS), squalene synthase (SQS), squalene monooxygenase or epoxidase (SQE), beta-amyrin synthase (BAS), and mixed function amyrin synthase (MFAS) genes were determined by qRT-PCR in different plant organs of R. officinalis, S. officinalis and T. persicus. The mRNA levels were normalized with reference to the housekeeping gene GAPDH. Error bars represent standard deviation from the mean (n = 3).
Based on the results obtained by ANOVA, BA, OA, UA and their total (TAs) content (mg/g DW) were statistically significant (P < 0.05) in different tissues and plant species, and in their interactions. In the plant species studied, the highest and the lowest content of TAs was observed in the leaf and root, respectively (Fig. 7a). R. officinalis showed the highest TAs concentration in all tissues except the root, while S. officinalis contained a higher content of TAs (7.50 ± 0.16) in the root in comparison with R. officinalis and T. persicus.
These results are consistent with those of an earlier report of TAs distribution in various plants, which introduced the Lamiaceae family as a particularly rich source of BA, OA, and UA, with the highest concentration measured in rosemary leaves24,26.
Regarding the studied TAs, the maximum content of BA was quantified in leaves of R. officinalis (11.20 ± 0.6 mg/g DW), while the highest content of OA (5.75 ± 0.52 mg/g DW) and UA (14.1 ± 0.98 mg/g DW) was determined in leaves of S. officinalis. The predominant TA in different tissues of R. officinalis was BA, and UA predominated in the studied tissues of S. officinalis and T. persicus. It has been proposed that the content of secondary metabolites can be significantly affected by genetics59,60, the plant species3,61 and organ62,63, and environmental conditions64,65, which is in accordance with the results obtained here. In contrast with the present study, in which BA was the predominant TA in R. officinalis, UA was the main TA observed in the study of Razboršek et al.26, whereas roughly equal proportions of BA, OA and UA were reported by Jäger et al.24.
The expression levels of the six genes of the TAs metabolic pathway were also determined by qRT-PCR in different plant tissues. The values expressed as means ± SD of three replicates were normalized with GAPDH as a reference gene (Fig. 7b). The gene expression levels were compared in two dimensions, cross-gene and cross-tissue. In the cross-gene comparison, the transcript level of a gene was compared with other genes in the same tissue, and in the cross-tissue comparison, the mRNA content of a gene in one tissue was compared with its expression in all other tissues. Significant differences were obtained in the results from the expression level analysis in each tissue and across tissues. Comparison of gene expression across tissues was used to infer the link between elevated expression and metabolite accumulation in tissues. The cross-tissue comparison found a variable expression of the genes in this pathway, with the highest level in leaves, followed by flowers and stems, and the lowest level in roots.
In the case of SQE, the same expression pattern was observed in all three plants, being distinctly tissue-specific, and the leaves showing by far the highest level. The expression of this gene was also more pronounced than that of other genes in other plants. The expression levels of RoSQE, SoSQE, and TpSQE were recorded as 5.96 ± 0.18, 4.39 ± 0.48 and 2.19 ± 0.18, respectively. Only in S. officinalis was the transcript level of SoBAS slightly higher compared to SoSQE (4.76 ± 0.45).
The expression level of OSC enzymes was consistent with TAs concentration in various tissues of the plants. High levels of expression were recorded for RoLUS (4.28 ± 0.24), SoBAS (4.76 ± 0.45) and TpBAS (1.56 ± 0.25) in leaves. The expression pattern of SoFDS varied significantly across the tissues, being strongly expressed in the stem (3.50 ± 0.46) and root (3.18 ± 0.67) and moderately expressed in leaf (2.92 ± 0.49) and flower (2.07 ± 0.32). The expression of the SQS gene was observed in all tissues, but the expression pattern of RoSQS, SoSQS, and TpSQS varied according to the tissues.
Correlation of TAs contents and gene expression
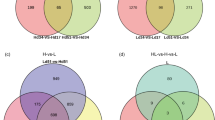
The correlation coefficient was calculated to indicate whether the tissue-specific changes in gene expression were directly associated with metabolite accumulation or not (Fig. 8). The amplitude of correlation between the TAs content and gene expression in different plant tissues varied among the plant species, but most genes had a positive correlation with TAs content. All correlations between TAs content and SQE gene expression were significant in R. officinalis, S. officinalis, and T. persicus and highly correlated with RoSQS, RoLUS, SoSQS, SoBAS and SoMFAS, whereas there was low or no correlation with TpLUS and TpSQS (Fig. 8). Considering the correlation coefficients between the genes of TAs biosynthetic pathway, SQE was highly correlated with the OSCs, which is dominant in each plant. For example, a strong association was observed between RoSQE and RoLUS.
Discussion
SMs are valuable compounds synthesized in plants by a series of chemical and multi-step reactions. In this enzymatic chain, the product of one reaction becomes the substrate for the following one. Therefore, the activity of each enzyme depends on the amount of substrate produced by the previous reaction. Rate-limiting steps in a metabolic pathway require the greatest activation energy and can alter the flux or metabolite concentration in the pathways. The rate of these reactions is influenced by enzyme activity rather than substrate concentration66. In this study, gene expression tracking was performed by transcriptome mining, co-expression network analysis, and tissue-specific metabolite-expression analysis to identify the key TAs biosynthetic genes that coordinately regulate the expression of other enzyme-encoding genes in Lamiaceae plants. First, putative key genes were extracted by direct analysis of the structure of the co-expression network. Several characteristics related to the network structure were calculated to provide insights into potential key genes. Highly connected genes in the network reflect relatively high information exchange with other nodes. Thus, this parameter, the relationship with the biological network, is considered as the key property for a gene.
Betweenness centrality (BC), another centrality measure frequently applied in diverse biological research, is defined as the number of shortest paths on which a node lies. “High traffic” nodes are those with large BC values67 and a strong impact on the network structure. In the genes with a high value, these two parameters may play an influential role in a given process. In the present study, four genes showed both hubs and nodes with a high betweenness (MVD1, AT3G54250, OSC and SQE). In gene co-expression networks, highly connected genes are more effective in the biological pathway. Nevertheless, the results of previous studies indicate that the last three enzymatic steps of the MVA pathway may not be important control points in plant terpene biosynthesis and are not correlated with the rate of terpene formation68. A weaker co-expression relationship is observed between this enzyme and downstream genes in the TAs pathway in spite of the high connectivity of MVD1 in the co-expression network. Thus, it is not a suitable target for metabolic engineering to maximize the yield of TAs.
Furthermore, it is recommended to focus on downstream reactions to understand which genes should be overexpressed to increase TAs production. From the co-expression network, it could be reasonably argued that SQE is probably the best choice in this regard, being responsible for increased substrate production or superior channeling of the substrate to the triterpene cyclases. Indeed, SQE overexpression has already been introduced as a new strategy for boosting triterpene production in plants69. Arabidopsis has six SQE copies in its genome69. Further, as shown in Fig. 3c, SQE has a co-expression relationship with OSCs, which points to OSCs as regulatory steps in the pathway orienting the biosynthetic flux towards TAs. Since highly co-expressed genes are more likely to be co-regulated, they display prominent connectivity patterns, which may play biologically influential or regulatory roles in a process. However, in several other reports, FDS and SQS were also co-expressed and used to increase the triterpene production70,71,72,73.
Metabolic changes are effected mainly through a single or a few enzymes, whose activity is most likely to lead to notable changes in metabolite concentrations. The activity of an enzyme may be related to various factors, including environmental conditions that may cause a change in gene expression. Therefore, the analysis of expression levels, metabolite concentration, and their connections has attracted considerable attention.
As depicted in Fig. 7, the TAs profile varied considerably across the tissues, showing almost the same pattern in the three Lamiaceae plants. The tissue specificity of several metabolic pathway genes was reflected in their expression level. Most of the genes associated with the TAs biosynthetic pathway were expressed mainly in the aerial parts of the plants. None of the studied genes are exclusively expressed in specific tissues, but they exhibited a spatial expression pattern (Fig. 7). Associations between metabolite accumulation and highly expressed genes were obvious in some cases. The most expressed genes in both cross-gene and cross-tissue dimensions are most likely to be involved in metabolite accumulation. For instance, SQE and some OSCs were highly expressed in leaves and presumably implicated in TAs concentrations, with a good correlation with metabolite accumulation. In contrast, FDS and SQS were expressed in all the studied tissues, and cross-tissue expression levels may give a narrow description with respect to the TAs concentrations.
The expression pattern of SoFDS was almost in line with previously obtained results from S. guaranitica and S. splendens, which exhibited the highest expression levels in the root74,75 and S. miltiorrhiza, in the stem76. However, our findings differ from those reported for S. officinalis, in which young leaves were the site of high expression of SoFDS77. Additionally, although FDS was expressed in all the studied tissues of the Lamiaceae plants, high levels of SoFDS were observed in the stem and root, and RoFDS and TpFDS in the leaf. The Lamiaceae family is rich in bioactive compounds, including phenols, alkaloids, monoterpene, sesquiterpenes, triterpenes, and steroids. Farnesyl diphosphate (FDP), produced by the key enzyme FDS, serves as a precursor for sesquiterpenoids, sterols, triterpenoids, polyprenols, side chains of ubiquinone, and polyisoprenoids such as natural rubber78,79. Based on GC-MS analysis of S. officinalis, sesquiterpenes formed the main group of compounds found in the stems. Since FDS enzyme activity is closely associated with the accumulation of subsequent products, the high expression level of the FDS gene in the stem may be related to the products originating from FDP.
The expression of the SQS gene was also observed in all tissues and the expression patterns of RoSQS, SoSQS and TpSQS were exactly matched in the transcriptome analysis of S. splendens (SRP126897), with the highest level of transcripts in the leaf 75. In general, 1–3 SQS genes are found in plants. In S. miltiorrhiza, two copies of SQS were reported80. Rong et al.81 confirmed that SmSQS2 is more expressed in the root than the leaf. Squalene is the precursor for the production of plant sterols as well as triterponoid backbones, so it is important for controlling the flux towards sterol vs. non-sterol products. Both triterpenoids and steroids have been found in the roots of some Salvia species82. In the plants studied here, a relatively high expression of SoSQS was observed in the root.
Although it should be taken into consideration that the nature of transcriptional activity is very dynamic, and can change rapidly with the climatic conditions, developmental stages, and environmental stresses, our findings point to potential key genes for targeting to boost TAs production. In this respect, we evaluated the correlation of specific transcript levels in tissues with BA, OA, and UA accumulation. TAs levels were found to be most significantly correlated with the expression of SQE in the three Lamiaceae plants (Fig. 8). In most cases, this gene was also highly correlated with OSCs, indicating a co-expression relationship, as observed in the co-expression network analysis. SQE is a rate-limiting enzyme in the biosynthetic pathway, oxidizing squalene into 2,3-oxidosqualene (squalene epoxide) by conducting a stereospecific epoxidation reaction83. In addition, it is considered as the biological precursor of all cyclic triterpenoids. Since a rate-limiting enzyme determines the overall rate of a metabolic pathway, SQE has a cascading control of the upregulation of the downstream genes84. Therefore, overexpression of this enzyme by metabolic engineering can constitute a system for producing high-value therapeutic TA compounds85.
The OSC enzyme family is responsible for a diversifying step in the biosynthesis of various triterpene/sterol scaffolds56 by cyclization of 2,3-oxidosqualene. Triterpenes/sterols are derived from the common substrate, 2,3-oxidosqualene, leading to the division of the 2,3-oxidosqualene substrate pool between sterols and a range of triterpenes. TAs scaffolds are derived via a chair–chair–chair (C-C-C) conformation dammarenyl cation by D-ring expansion to form lupeol or further E-ring expansion to form β-amyrin and α-amyrin86. These compounds are produced as sole products by the LUS, BAS, and MFAS enzymes, respectively.
OSCs with the same functions exhibit a high sequence identity regardless of plant species. Mutagenesis experiments revealed that the functional diversity of OSCs is determined by certain key amino acid residues, which are essential for the subsequent product specificity of the cyclization process87. The end products BA, OA, and UA are produced by further modification by tailoring enzymes (P450s). Additionally, metabolic fluxes can shift the precursor reservoir in the direction of a preferred compound to improve TAs yields. Experimental evidence confirmed that the suppression of a competitive branch may lead to the diversion of the substrate pool toward other OSCs and increase the desired metabolite production88. This goal requires the metabolic branch point genes and the upstream genes to be sufficiently flexible to respond to induced genetic and environmental changes such as elicitor application. In the present study, the flexibility of these genes was confirmed by transcriptome mining, which showed that the elicitors YE, MeJA and salicylic acid (SA) caused up-regulation of the OSCs or differential transcriptional regulation of each OSC in different organs of the plant. The results of various studies confirm that these elicitors can induce de novo synthesis of SMs in plants through transcriptional alteration of the genes in the biosynthetic pathway89.
Several studies focusing on the regulatory function of FDS and SQS in sesquiterpene, phytosterol, cholesterol, and triterpene saponin biosynthesis42,70,72,90,91 report that the activity of these enzymes is associated with the overall yield of these SMs. As illustrated in Fig. 7, the leaves in the studied Lamiaceae plants showed high expression levels of enzymes and metabolite concentrations. In general, striking variability was found among the tissue-specific expression profiles of TAs enzymes. These results indicate that TAs accumulation may be connected with the expression of the genes encoding each of the six enzymes. In addition, the existence of weak or negative correlations between TAs content and the expression of TpSQS and SoFDS genes does not necessarily mean a negative or no relationship, as in many cases it may be related to a change in the metabolic flow towards non-triterpenic compounds in a specific tissue. Thus, the correlation may be high despite the lower transcript levels in the tissue. Furthermore, the transcriptional level is not always directly related to the enzymatic activity, since several regulatory systems are involved in the availability and activity of the final protein92.
Since high-value SMs are produced by plants in low amounts, an in-depth understanding of their biosynthetic machinery is difficult. To increase these compounds, which often have pharmaceutical properties, metabolic engineering necessitates a comprehensive knowledge of metabolic pathways and fluxes, and how these change in response to various types of genetic and environmental perturbations. Other factors that need to be understood include enzyme activity, kinetic properties, post-processing, and all regulatory systems. These factors are critical for elucidating the control of metabolic flux, the desired genetic manipulation, and biotechnological applications.
In the present work, the in silico analysis of gene expression and qRT-PCR provided information about the expression behavior of selected genes of the TAs metabolic pathway in the Lamiaceae plants. These results can be considered as a valuable step toward a rational selection of genes suitable for manipulation to improve TAs production. The results of the network analysis were confirmed by the correlation of TAs contents and gene expression. SQE and OSC expression levels were consistent with the TAs concentration in various plant tissues. An adequate combination of statistical methods and network structure analysis can lead to a significant improvement in our knowledge of the underlying biological functions of a set of genes. Furthermore, this approach can link the structure of the network to a biological process and identify key genes. Further investigation is necessary to identify other factors potentially involved in the regulatory process.
Methods
Plant material and data collection
Different plant parts, namely leaf, stem, flower, and root, were collected from three members of the mint family: Rosmarinus officinalis L., Salvia officinalis L. and Thymus persicus (Ronniger ex Rech. f.) Jalas cultivated in the research field of the Medicinal Plants and Drugs Research Institute (MPDRI), Shahid Beheshti University, Evin, Tehran (35° 48′ N, 51° 23′ E at an altitude of 1800 m), Iran. After collection, plant materials were immediately frozen in liquid nitrogen for subsequent metabolic analysis and RNA extraction. A voucher specimen of T. persicus (MPH-1673), S. officinalis (MPH-2638), and R. officinalis (MPH-2639) has been deposited at the herbarium of MPDRI, Shahid Beheshti University (MPH).
Respective datasets were extracted from the Sequence Read Archive (SRA) database of NCBI (https://www.ncbi.nlm.nih.gov/sra/), with accession numbers SRP006813, SRP065934 SRP126897 and, SRP111399. The transcriptome data of SRP006813 included eighteen samples of R. officinalis, which were submitted by Michigan State University. Among all the data, six different tissue samples were selected for in-silico expression analysis. Assembled transcripts of R. officinalis were downloaded from the Medicinal Plant Genomics Resource (http://medicinalplantgenomics.msu.edu/). SRP126897 was associated with the project of RNA-Seq of S. splendens (red and purple flowers) with 30 samples of different organs75. SRP065934 consisted of eight samples of transcriptional profiles related to S. miltiorrhiza cell cultures in response to salicylic acid (SA) elicitation93. In addition, SRP111399 was related to five samples of expression profiling by high throughput sequencing of S. miltiorrhiza after elicitation by methyl jasmonate (MeJA) and yeast extract (YE)94.
Data preprocessing and differentially expressed gene (DEG) screening
SRA Toolkit version 2.8.2 (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) was used to convert the SRA files to FASTQ format. Before any analysis, the quality of raw sequence reads was assessed by FASTQC software. In order to obtain clean reads, Trimmomatic program, Version. 0.36 (http://www.usadellab.org/cms/?page = trimmomatic)95 was used to trim low-quality bases from both ends of raw Illumina reads and adapter removal. After trimming, de novo assembly was performed using Trinity v2.6.696 at the default KMER setting for the datasets without available assembly files. In the next procedure, the expression levels of the re-constructed transcripts in the individual libraries (different tissues and elicitations) were measured by mapping high-quality filtered reads to assembled transcriptomes using Bowtie-2 via RSEM with recommended RSEM parameters. RSEM uses SAM tool libraries for sorting and indexing the mapping results. “R” and the Bioconductor package edge R were used to analyze the expected counts produced by RSEM. In differential expression analysis of any of the two sample groups, p-values less than 0.05 were considered as the significance threshold and genes with positive and negative log fold change values (logFC) ≥ │2│ were described as up- or downregulated genes, respectively.
Functional annotation and scrutiny of the expression pattern of the genes involved in TAs biosynthetic pathway
In order to validate and annotate the assembled unigenes, the searches for sequence similarity were performed using the BLAST algorithm against the NCBI non-redundant (nr), SWISS-PROT protein, NCBI nucleotide (nt), and KEGG with the e-value cut-off of 1e-5. In addition, heatmap matrices were used for pattern recognition and visualization to identify expression pattern of the TAs biosynthetic genes in each experiment. Then, all the statistical analysis and plots were conducted in an R environment (version 5.3.0). Further, the co-expression network was constructed in Cytoscape v 3.6.1, an open source software platform, by the GeneMANIA Cytoscape plugin97 based on the genes of the TAs biosynthetic pathway. The plugin uses a large database of functional interaction networks from multiple organisms including Arabidopsis thaliana. Accordingly, the network properties including specific centrality parameters were calculated by the CentiScaPe2 plugin to identify key nodes and edges in the target biosynthetic pathway.
Bioinformatics analysis for primer design
The predicted protein sequences of the targeted TAs biosynthetic enzymes in R. officinalis were subjected to phylogenetic analysis with the proteins of other plants. The protein sequences of these enzymes were then retrieved from NCBI GenBank databases (http://www.ncbi.nlm.nih.gov/). The amino acid sequences were aligned using ClustalW with default parameters as implemented in the program MEGA7 and the phylogenetic tree was built using the Neighbor-Joining Method. In addition, pairwise genetic distances between the sequences were calculated and the closely related plant species were selected based on the genetic distance matrix for further analysis. Motif-based sequence analysis tools, Multiple Expectation – Maximization for Motif Elicitation (MEME) Suite 5.0.0, (http://meme-suite.org/tools/meme)98 were used to discover novel and ungapped motifs in the protein sequences. The parameters of MEME analyses were applied: the distribution of motif occurrences, zero or one per sequence, the number of different motifs (20), minimum motif width (6); and maximum motif width (10). All possible pairs of motifs were detected to design taxa-specific degenerate primers based on conserved regions, and the best primers were selected for the PCR analysis. A Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to increase the chance of finding specific primer pairs. The tool can incorporate a global alignment mechanism and is designed to be very sensitive in detecting potential amplification targets. The properties of each primer such as melting temperature, GC content percentage, and polymerase chain reaction (PCR) suitability were checked with some online tools like OligoCalc (http://biotools.nubic.northwestern.edu/OligoCalc.html) and OligoAnalyzer (https://eu.idtdna.com/calc/analyzer).
RNA isolation, RT reaction, PCR amplification and sequencing
Total RNA was extracted using Trizol (TRI reagent Sigma-Aldrich, USA), and the first-strand cDNA was synthesized from 2 µg of the isolated RNA templates by reverse transcriptase with Oligo-(dT)18 primers according to the instructions of the RevertAidTM H M-MuLV First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Fermentas). PCR was performed using the cDNA prepared from the leaves of the studied plants as templates to amplify DNA fragments encoding TAs biosynthetic enzymes. Each PCR reaction was set up in 25 μl volume containing 12.5 μl 2X PCR master mix (Sinaclon, Iran), 50 ng of the synthesized cDNA and gene-specific primers (Table 1). The PCR conditions included initial denaturation at 94 °C for 5 min, followed by 35 cycles [94 °C for 1 min, 50–55 °C (depending on the primer combination) for 30 seconds, 72 °C for 30 sec] and a final extension at 72 °C for 5 min. The amplified products were separated on 1.5% agarose gel and visualized by ethidium bromide staining. Purification and concentration of the PCR product were carried out using the GF-1 Gel DNA Recovery Kit (Vivantis). Purified PCR products were sequenced and screened in the GenBank (NCBI) database for matching homology. First, the amplicon sequences were aligned by applying BLASTx against protein databases, including the non-redundant (Nr) protein database, Swiss-Prot (http://www.expasy.ch/sprot/), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.jp/kegg/), and then using BLASTn against the Nucleotide (Nt) database (with a threshold of E < 1.0E-5).
Transcript levels of the genes involved in the biosynthesis of TAs
qPCR was done to investigate the transcript levels of six genes encoding TAs biosynthetic enzymes in different organs of R. officinalis, S. officinalis and T. persicus. The qPCR reactions were performed in a 20 μl volume including 10 μl qPCR GreenMaster (Jena Bioscience, Germany), 50 ng of cDNA and 300 nM of each primer in a Rotor-Gene® 6000 (Qiagen, Germany). PCR amplification was performed with three technical replications under the following conditions: 2 min at 95 °C, followed by 45 cycles of 95 °C for 20 s, 53 °C for 20 s, and 72 °C for 20 s. Then, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used to normalize the real-time PCR data as an internal control. The expression level was calculated according to the 2−ΔΔCt method (Livak and Schmittgen, 2001) based on the mean of three independent determinations of the threshold cycle. In addition, the value of 2−ΔΔCt was used to represent the relative expression of each gene.
Extraction and HPLC analysis
In the next procedure, reverse phase-HPLC was used to determine the content of BA, OA, and UA in the studied plant parts. As mentioned, lyophilized and powdered plant material (1.0 g) was drenched in MeOH (40 mL) and immediately sonicated at 30% amplitude for 40 min at room temperature99,100. The analysis was conducted using a Water symmetry C18 column with methanol: phosphoric acid: water (87:0.05:12.95) as an isocratic elution mode and UV detection (λ = 210 nm). All standards of BA, OA, and UA were purchased from Sigma-Aldrich and the standard solutions were prepared by serial dilutions of stock solutions at a concentration range of 50–800 ppm. A calibration curve (standard curve) was generated by injecting the series of calibration standards and plotting concentration against the peak area in Excel 2016. Then, the three TAs in the unknown samples were quantified by the standard equation obtained from the calibration curve, and the correlation coefficients (R2) were calculated for each of the three compounds. Each sample was run three times to evaluate the precision.
Statistical analysis
All data are expressed as means ± SD. Normality was assessed using the Shapiro-Wilk test and the data were analyzed by univariate one-way ANOVA. Duncan’s multiple range test was used for post hoc analysis. To measure the intensity of correlation between the expression of each gene and TAs content, the Pearson correlation coefficient was calculated for different organs of each plant and represented in correlograms. Analyses were performed by R statistical software v.3.5.0 (freely available at http://www.r-project.org). In all the analyses, the P < 0.05 was used as the significance threshold.
References
Hill, R. A. & Connolly, J. D. Triterpenoids. Nat. Prod. Rep 30, 1028–1065 (2013).
Janicsák, G., Veres, K., Kakasy, A. Z. & Máthé, I. Study of the oleanolic and ursolic acid contents of some species of the Lamiaceae. Biochem. Syst. Ecol. 34, 392–396 (2006).
Mirjalili, M., Ayyari, M., Bakhtiar, Z., Moridi Farimani, M. & Sonboli, A. Quantification of betulinic, oleanolic and ursolic acids as medicinally important triterpenoids in some Thymus species from Iran. Res. J. Pharmacogn. 3, 23–28 (2016).
Pai, S. R., Upadhya, V., Hegde, H. V., Joshi, R. K. & Kholkute, S. D. Determination of betulinic acid, oleanolic acid and ursolic acid from Achyranthes aspera L. using RP-UFLC-DAD analysis and evaluation of various parameters for their optimum yield. Indian J Exp Biol. 54, 196–120 (2016).
Laszczyk, M. N. Pentacyclic triterpenes of the lupane, oleanane and ursane group as tools in cancer therapy. Planta Med. 75, 1549–1560 (2009).
Cassels, B. K. & Asencio, M. Anti-HIV activity of natural triterpenoids and hemisynthetic derivatives 2004–2009. Phytochem. Rev. 10, 545–564 (2011).
Peláez, G. L. M., Sierra, J. A., Alzate, F., Holzgrabe, U. & Ramirez-Pineda, J. R. Pentacyclic triterpenes from Cecropia telenitida with immunomodulatory activity on dendritic cells. Rev. bras. farmacogn. 23, 754–761 (2013).
Shai, L., McGaw, L., Aderogba, M., Mdee, L. & Eloff, J. Four pentacyclic triterpenoids with antifungal and antibacterial activity from Curtisia dentata (Burm. f) CA Sm. leaves. J Ethnopharmacol 119, 238–244 (2008).
Gai, W. T., Yu, D. P., Wang, X. S. & Wang, P. T. Anti-cancer effect of ursolic acid activates apoptosis through ROCK/PTEN mediated mitochondrial translocation of cofilin-1 in prostate cancer. Oncol Lett 12, 2880–2885 (2016).
Lewinska, A., Bednarz, D., Adamczyk-Grochala, J. & Wnuk, M. Phytochemical-induced nucleolar stress results in the inhibition of breast cancer cell proliferation. Redox Biol 12, 469–482 (2017).
Potze, L., Mullauer, F., Colak, S., Kessler, J. & Medema, J. Betulinic acid-induced mitochondria-dependent cell death is counterbalanced by an autophagic salvage response. Cell Death Dis 5, e1169 (2014).
Chudzik, M., Korzonek-Szlacheta, I. & Król, W. Triterpenes as potentially cytotoxic compounds. Molecules 20, 1610–1625 (2015).
Kikuchi, T. et al. Cytotoxic and apoptosis-inducing activities of triterpene acids from Poria cocos. J. Nat. Prod. 74, 137–144 (2011).
Mukherjee, R., Kumar, V., Srivastava, S. K., Agarwal, S. K. & Burman, A. C. Betulinic acid derivatives as anticancer agents: structure activity relationship. Anticancer Agents Med Chem 6, 271–279 (2006).
Zhang, W., Men, X. & Lei, P. Review on anti-tumor effect of triterpene acid compounds. J. Cancer Res. Ther. 10, 14 (2014).
Zhu, Y. Y., Huang, H. Y. & Wu, Y. L. Anticancer and apoptotic activities of oleanolic acid are mediated through cell cycle arrest and disruption of mitochondrial membrane potential in HepG2 human hepatocellular carcinoma cells. Mol Med Rep 12, 5012–5018 (2015).
Silva, F. S., Oliveira, P. J. & Duarte, M. F. Oleanolic, ursolic, and betulinic acids as food supplements or pharmaceutical agents for type 2 diabetes: promise or illusion? J. Agric. Food Chem. 64, 2991–3008 (2016).
Oliveira, B. H. d., Santos, C. A. & Espíndola, A. P. D. Determination of the triterpenoid, betulinic acid, in Doliocarpus schottianus by HPLC. Phytochem. Anal. 13, 95–98 (2002).
Neto, C. C. Cranberry and its phytochemicals: a review of in vitro anticancer studies. J. Nutr. 137, 186S–193S (2007).
Strzemski, M. et al. Carlina species as a new source of bioactive pentacyclic triterpenes. Ind Crops Prod 94, 498–504 (2016).
Ayatollahi, A. M. et al. Pentacyclic triterpenes in Euphorbia microsciadia with their T-cell proliferation activity. Iran J Pharm Res 10, 287 (2011).
Khan, S., Kazmi, M. H., Ahmed, E., Malik, A. & Sharif, A. Phytochemical Studies on Sorbus cashmiriana. J. Chem. Soc. Pak. 35, 130–134 (2013).
Begum, S., Siddiqui, S., Khatoon, R. & Aftab, F. Phytochemical studies on Syzygium aromaticum Linn. J. Chem. Soc. Pak 36, 512 (2014).
Jäger, S., Trojan, H., Kopp, T., Laszczyk, M. N. & Scheffler, A. Pentacyclic triterpene distribution in various plants–rich sources for a new group of multi-potent plant extracts. Molecules 14, 2016–2031 (2009).
Hedayati, A., Mirjalili, M. H. & Hadian, J. Quantification of betulinic, oleanolic and ursolic acid as medicinally important triterpenoids in wild and in vitro callus culture of Salvia sahendica (Lamiaceae): a comparative study. J Bio Env Sci 6, 327–332 (2015).
Razboršek, M. I., Vončina, D. B., Doleček, V. & Vončina, E. Determination of oleanolic, betulinic and ursolic acid in Lamiaceae and mass spectral fragmentation of their trimethylsilylated derivatives. Chromatographia 67, 433–440 (2008).
Topçu, G. Bioactive triterpenoids from Salvia species. J. Nat. Prod. 69, 482–487 (2006).
Bakhtiar, Z., Mirjalili, M. H., Sonboli, A., Farimani, M. M. & Ayyari, M. In vitro propagation, genetic and phytochemical assessment of Thymus persicus—a medicinally important source of pentacyclic triterpenoids. Biologia 69, 594–603 (2014).
Kim, H.-I. et al. Inhibition of estrogen signaling through depletion of estrogen receptor alpha by ursolic acid and betulinic acid from Prunella vulgaris var. lilacina. Biochem. Biophys. Res. Commun. 451, 282–287 (2014).
Hossain, M. A. & Ismail, Z. Isolation and characterization of triterpenes from the leaves of Orthosiphon stamineus. ARAB J CHEM 6, 295–298 (2013).
Oldfield, E. & Lin, F. Y. Terpene biosynthesis: modularity rules. Angew. Chem. Int. Ed. Engl. 51, 1124–1137 (2012).
Sawai, S. & Saito, K. Triterpenoid biosynthesis and engineering in plants. Front Plant Sci 2, 25 (2011).
Thimmappa, R., Geisler, K., Louveau, T., O’Maille, P. & Osbourn, A. Triterpene biosynthesis in plants. Annu Rev Plant Biol 65, 225–257 (2014).
Bradley, P. H., Brauer, M. J., Rabinowitz, J. D. & Troyanskaya, O. G. Coordinated concentration changes of transcripts and metabolites in Saccharomyces cerevisiae. Plos Comput. Biol. 5, e1000270 (2009).
Patil, K. R. & Nielsen, J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl. Acad. Sci. USA 102, 2685–2689 (2005).
Urbanczyk-Wochniak, E. et al. Parallel analysis of transcript and metabolic profiles: a new approach in systems biology. EMBO Rep. 4, 989–993 (2003).
Zelezniak, A., Sheridan, S. & Patil, K. R. Contribution of network connectivity in determining the relationship between gene expression and metabolite concentration changes. Plos Comput. Biol. 10, e1003572 (2014).
Carrera, J., Rodrigo, G., Jaramillo, A. & Elena, S. F. Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 10, R96 (2009).
Windram, O., Penfold, C. A. & Denby, K. J. Network modeling to understand plant immunity. Annu Rev Phytopathol 52 (2014).
Van Dam, S., Võsa, U., van der Graaf, A., Franke, L. & de Magalhães, J. P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinformatics, bbw139 (2017).
Ren, W. et al. Combining multi-dimensional data to identify key genes and pathways in gastric cancer. PeerJ 5, e3385 (2017).
Yang, Y., Ge, F., Sun, Y., Liu, D. & Chen, C. Strengthening triterpene saponins biosynthesis by over-expression of farnesyl pyrophosphate synthase gene and RNA interference of cycloartenol synthase gene in Panax notoginseng cells. Molecules 22, 581 (2017).
Zhang, C. & Sun, Q. Weighted gene co-expression network analysis of gene modules for the prognosis of esophageal cancer. J. Huazhong Univ. Sci. Technol. Med. Sci. 37, 319–325 (2017).
Weßling, R. et al. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375 (2014).
Zhang, J. et al. Coexpression network analysis of the genes regulated by two types of resistance responses to powdery mildew in wheat. Sci. Rep. 6, 23805 (2016).
Palumbo, M. C. et al. Integrated network analysis identifies fight-club nodes as a class of hubs encompassing key putative switch genes that induce major transcriptome reprogramming during grapevine development. Plant Cell 26, 4617–4635 (2014).
Sun, W. et al. Weighted gene co-expression network analysis of the dioscin rich medicinal plant Dioscorea nipponica. Front Plant Sci 8, 789 (2017).
Yu, Y. et al. Transcriptome analysis reveals key differentially expressed genes involved in wheat grain development. Crop J 4, 92–106 (2016).
Amrine, K. C., Blanco-Ulate, B. & Cantu, D. Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PLoS One 10, e0118731 (2015).
Krishnan, A., Gupta, C., Ambavaram, M. M. & Pereira, A. RECoN: Rice Environment Coexpression Network for Systems Level Analysis of Abiotic-Stress Response. Front Plant Sci 8, 1640 (2017).
Lorenz, W. W. et al. Microarray analysis and scale-free gene networks identify candidate regulators in drought-stressed roots of loblolly pine (P. taeda L.). BMC Genomics 12, 264 (2011).
Wei, X., Kuhn, D. N. & Narasimhan, G. In Bioinformatics Conference, 2003. CSB 2003. Proceedings of the 2003 IEEE. 75–83 (IEEE).
Zuiter, A. S., Sawwan, J. & Al Abdallat, A. Designing universal primers for the isolation of DNA sequences encoding Proanthocyanidins biosynthetic enzymes in Crataegus aronia. BMC Res Notes 5, 427 (2012).
Kim, Y. S. et al. Gene regulation patterns in triterpene biosynthetic pathway driven by overexpression of squalene synthase and methyl jasmonate elicitation in Bupleurum falcatum. Planta 233, 343–355 (2011).
Lu, X., Tang, K. & Li, P. Plant metabolic engineering strategies for the production of pharmaceutical terpenoids. Front Plant Sci 7, 1647 (2016).
Misra, R. C., Maiti, P., Chanotiya, C. S., Shanker, K. & Ghosh, S. Methyl jasmonate-elicited transcriptional responses and pentacyclic triterpene biosynthesis in sweet basil. Plant Physiol., pp. 113.232884 (2013).
Naoumkina, M. A. et al. Genomic and coexpression analyses predict multiple genes involved in triterpene saponin biosynthesis in Medicago truncatula. Plant Cell 22, 850–866 (2010).
Zeng, H. et al. Comparative analysis of the major chemical constituents in Salvia miltiorrhiza roots, stems, leaves and flowers during different growth periods by UPLC-TQ-MS/MS and HPLC-ELSD methods. Molecules 22, 771 (2017).
Hadian, J., Hossein Mirjalili, M., Reza Kanani, M., Salehnia, A. & Ganjipoor, P. Phytochemical and morphological characterization of Satureja khuzistanica Jamzad populations from Iran. Chem. Biodivers. 8, 902–915 (2011).
Selseleh, M. et al. Metabolic diversity and genetic association between wild populations of Verbascum songaricum (Scrophulariaceae). Ind Crops Prod 137, 112–125 (2019).
Abdollahi-Ghehi, H., Sonboli, A., Ebrahimi, S. N., Esmaeili, M. A. & Mirjalili, M. H. Triterpenic Acid Content and Cytotoxicity of Some Salvia Species From Iran. Nat Prod Commun 14, 1934578X19842722 (2019).
Butkiene, R., Budiene, J. & Judzentiene, A. Variation of secondary metabolites (essential oils) in various plant organs of Juniperus communis L. wild growing in Lithuania. Balt. For. 21, 59–64 (2015).
Raeisi, M., Tajik, H., Aliakbarlu, J., Mirhosseini, S. H. & Hosseini, S. M. H. Effect of carboxymethyl cellulose-based coatings incorporated with Zataria multiflora Boiss. essential oil and grape seed extract on the shelf life of rainbow trout fillets. Lebensm. Wiss. Technol. 64, 898–904 (2015).
Khonakdari, M. R., Mirjalili, M. H., Gholipour, A., Rezadoost, H. & Farimani, M. M. Quantification of galantamine in Narcissus tazetta and Galanthus nivalis (Amaryllidaceae) populations growing wild in Iran. Plant Genet. Resour. Newsl. 16, 188–192 (2018).
Pourhosseini, S. H., Hadian, J., Sonboli, A., Nejad Ebrahimi, S. & Mirjalili, M. H. Genetic and Chemical Diversity in Perovskia abrotanoides Kar.(Lamiaceae) Populations Based on ISSR s Markers and Essential Oils Profile. Chem. Biodivers. 15, e1700508 (2018).
Morandini, P. Control limits for accumulation of plant metabolites: brute force is no substitute for understanding. Plant Biotechnol. J. 11, 253–267 (2013).
Azuaje, F. J. Selecting biologically informative genes in co-expression networks with a centrality score. Biol. Direct 9, 12 (2014).
Tholl, D. & Lee, S. Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book 9 (2011).
Dong, L. et al. Co-expression of squalene epoxidases with triterpene cyclases boosts production of triterpenoids in plants and yeast. Metab. Eng. 49, 1–12 (2018).
Kim, O. T. et al. Upregulation of phytosterol and triterpene biosynthesis in Centella asiatica hairy roots overexpressed ginseng farnesyl diphosphate synthase. Plant Cell Rep. 29, 403–411 (2010).
Kim, Y.-K. et al. Enhanced triterpene accumulation in Panax ginseng hairy roots overexpressing mevalonate-5-pyrophosphate decarboxylase and farnesyl pyrophosphate synthase. ACS Synth. Biol. 3, 773–779 (2014).
Lee, M.-H. et al. Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 45, 976–984 (2004).
Wu, S. et al. Engineering triterpene metabolism in tobacco. Planta 236, 867–877 (2012).
Ali, M. et al. De novo transcriptome sequencing and metabolite profiling analyses reveal the complex metabolic genes involved in the terpenoid biosynthesis in Blue Anise Sage (Salvia guaranitica L.). DNA Res (2018).
Ge, X., Chen, H., Wang, H., Shi, A. & Liu, K. De novo assembly and annotation of Salvia splendens transcriptome using the Illumina platform. PLoS One 9, e87693 (2014).
Ma, X.-H. et al. The biosynthetic pathways of tanshinones and phenolic acids in Salvia miltiorrhiza. Molecules 20, 16235–16254 (2015).
Ali, M. et al. Transcriptome and metabolite analyses reveal the complex metabolic genes involved in volatile terpenoid biosynthesis in garden sage (Salvia officinalis). Sci. Rep. 7, 16074 (2017).
Dhar, M. K., Koul, A. & Kaul, S. Farnesyl pyrophosphate synthase: a key enzyme in isoprenoid biosynthetic pathway and potential molecular target for drug development. N Biotechnol 30, 114–123 (2013).
Sando, T. et al. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 72, 2049–2060 (2008).
Ma, Y., Yuan, L., Zhang, L., Hou, X. & Lu, S. Cloning and identification of two squalene synthase genes from Salvia miltiorrhiza. Chin. Tradit. Herb. Drugs 45, 1307–1312 (2014).
Rong, Q. et al. Molecular cloning and functional analysis of squalene synthase 2 (SQS2) in Salvia miltiorrhiza Bunge. Front Plant Sci 7, 1274 (2016).
Kolak, U., TOPÇU, G., Birteksöz, S., Ötük, G. & Ulubelen, A. Terpenoids and steroids from the roots of Salvia blepharochlaena. TURK J CHEM 29, 177–186 (2005).
He, F., Zhu, Y., He, M. & Zhang, Y. Molecular cloning and characterization of the gene encoding squalene epoxidase in Panax notoginseng. DNA Seq. 19, 270–273 (2008).
Han, J.-Y., In, J.-G., Kwon, Y.-S. & Choi, Y.-E. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry 71, 36–46 (2010).
Takemura, T., Chow, Y.-l., Todokoro, T., Okamoto, T. & Sato, F. In Plant Secondary Metabolism Engineering 95–109 (Springer, 2010).
Xu, R., Fazio, G. C. & Matsuda, S. P. On the origins of triterpenoid skeletal diversity. Phytochemistry 65, 261–291 (2004).
Xue, Z. et al. Identification of key amino acid residues determining product specificity of 2, 3-oxidosqualene cyclase in Oryza species. New Phytol. 218, 1076–1088 (2018).
Dhar, N. et al. Cloning and functional characterization of three branch point oxidosqualene cyclases from Withania somnifera (L.) dunal. J. Biol. Chem., jbc. M114, 571919 (2014).
Sailo, L. et al. In Biotechnological Approaches for Medicinal and Aromatic Plants 63–86 (Springer, 2018).
Bhat, W. W. et al. Molecular cloning, bacterial expression and promoter analysis of squalene synthase from Withania somnifera (L.) Dunal. Gene 499, 25–36 (2012).
Chen, D.-H., Ye, H.-C. & Li, G.-F. Expression of a chimeric farnesyl diphosphate synthase gene in Artemisia annua L. transgenic plants via Agrobacterium tumefaciens-mediated transformation. Plant Sci. 155, 179–185 (2000).
Vogel, C. & Marcotte, E. M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227 (2012).
Zhang, X. et al. Transcriptome sequencing in response to salicylic acid in Salvia miltiorrhiza. PLoS One 11, e0147849 (2016).
Zhou, W. et al. Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep. 7, 10554 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644 (2011).
Montojo, J. et al. GeneMANIA Cytoscape plugin: fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 (2010).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Srivastava, P. & Chaturvedi, R. Simultaneous determination and quantification of three pentacyclic triterpenoids—betulinic acid, oleanolic acid, and ursolic acid—in cell cultures of Lantana camara L. In Vitro Cell. Dev. Biol., Plant 46, 549–557 (2010).
Wang, H., Wang, Z. & Guo, W. Comparative determination of ursolic acid and oleanolic acid of Macrocarpium officinalis (Sieb. et Zucc.) Nakai by RP-HPLC. Ind Crops Prod 28, 328–332 (2008).
Acknowledgements
This research was supported by Shahid Beheshti University Research Council and University of Guilan, Iran. We thank Prof. C. Robin Buell (Department of Plant Biology of Michigan State University, USA) for her kind guidance and providing the data and assembly files of R. officinalis. We also thank Prof. Javier Palazón (University of Barcelona, Spain) for his kind help in proof reading and language editing of the manuscript.
Author information
Authors and Affiliations
Contributions
All Authors made equal contribution to the planning and development of experiments. Z.A., and M.T. performed the experiment and analyzed the data. M.H.M. contributed most to the phytochemical aspects. Z.A. wrote the manuscript. B.R. and M.H.M. revised the manuscript. All authors reviewed and approved the final version of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Aminfar, Z., Rabiei, B., Tohidfar, M. et al. Identification of key genes involved in the biosynthesis of triterpenic acids in the mint family. Sci Rep 9, 15826 (2019). https://doi.org/10.1038/s41598-019-52090-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52090-z
This article is cited by
-
RNA-seq analysis reveals narrow differential gene expression in MEP and MVA pathways responsible for phytochemical divergence in extreme genotypes of Thymus daenensis Celak
BMC Genomics (2024)
-
Characterization of essential oil profiles, triterpenic acids, and biological assay in aerial parts of various Thymus persicus Jalas (Ronniger ex Rech.f.) populations
Chemical and Biological Technologies in Agriculture (2023)
-
An insight into microRNA biogenesis and its regulatory role in plant secondary metabolism
Plant Cell Reports (2022)
-
Comparative transcriptome mining for terpenoid biosynthetic pathway genes in wild and cultivated species of Plantago
Protoplasma (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.