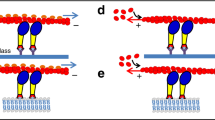
Abstract
Cells sense the rigidity of their environment through localized pinching, which occurs when myosin molecular motors generate contractions within actin filaments anchoring the cell to its surroundings. We present high-resolution experiments performed on these elementary contractile units in cells. Our experimental results challenge the current understanding of molecular motor force generation. Surprisingly, bipolar myosin filaments generate much larger forces per motor than measured in single-molecule experiments. Furthermore, contraction to a fixed distance, followed by relaxation at the same rate, is observed over a wide range of matrix rigidities. Finally, stepwise displacements of the matrix contacts are apparent during both contraction and relaxation. Building on a generic two-state model of molecular motor collections, we interpret these unexpected observations as spontaneously emerging features of a collective motor behaviour. Our approach explains why, in the cellular context, collections of resilient and slow motors contract in a stepwise fashion while collections of weak and fast motors do not. We thus rationalize the specificity of motor contractions implied in rigidity sensing compared to previous in vitro observations.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding authors upon request.
References
Moore, S. W., Roca-Cusachs, P. & Sheetz, M. P. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev. Cell 19, 194–206 (2010).
Wolfenson, H., Iskratsch, T. & Sheetz, M. P. Early events in cell spreading as a model for quantitative analysis of biomechanical events. Biophys. J. 107, 2508–2514 (2014).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Schiller, H. B. et al. β1- and αv-class integrins cooperate to regulate myosin II during rigidity sensing of fibronectin-based microenvironments. Nat. Cell Biol. 15, 625–636 (2013).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Levental, K. R. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Engler, J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Wolfenson, H. et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat. Cell Biol. 18, 33–42 (2016).
Howard, J. Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, 2001).
Kaya, M., Tani, Y., Washio, T., Hisada, T. & Higuchi, H. Coordinated force generation of skeletal myosins in myofilaments through motor coupling. Nat. Commun. 8, 16036 (2017).
Yasuda, K., Shindo, Y. & Ishiwata, S. Synchronous behavior of spontaneous oscillations of sarcomeres in skeletal myofibrils under isotonic conditions. Biophys. J. 70, 1823–1829 (1996).
Ajdari, A., Prost, J. & Jülicher, F. Modeling molecular motors. Rev. Mod. Phys. 69, 1269–1281 (1997).
Jülicher, F. & Prost, J. Spontaneous oscillations of collective molecular motors. Phys. Rev. Lett. 78, 4510–4513 (1997).
Plaçais, P. Y., Balland, M., Guérin, T., Joanny, J. F. & Martin, P. Spontaneous oscillations of a minimal actomyosin system under elastic loading. Phys. Rev. Lett. 103, 158102 (2009).
Camalet, S., Duke, T., Jülicher, F. & Prost, J. Auditory sensitivity provided by self-tuned critical oscillations of hair cells. Proc. Natl Acad. Sci. USA 97, 3183–3188 (2000).
Guérin, T., Prost, J. & Joanny, J. F. Motion reversal of molecular motor assemblies due to weak noise. Phys. Rev. Lett. 106, 068101 (2011).
Giannone, G. et al. Periodic lamellipodial contractions correlate with rearward actin waves. Cell 116, 431–443 (2004).
Giannone, G. et al. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 128, 561–575 (2007).
Ghassemi, S. et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc. Natl Acad. Sci. USA 109, 5328–5333 (2012).
Yang, B. et al. Mechanosensing controlled directly by tyrosine kinases. Nano Lett. 16, 5951–5961 (2016).
Liu, X. & Pollack, G. H. Stepwise sliding of single actin and myosin filaments. Biophys. J. 86, 353–358 (2004).
Stam, S., Alberts, J., Gardel, M. L. & Munro, E. Isoforms confer characteristic force generation and mechanosensation by myosin II filaments. Biophys. J. 108, 1997–2006 (2015).
Thoresen, T., Lenz, M. & Gardel, M. L. Thick filament length and isoform composition determine self-organized contractile units in actomyosin bundles. Biophys. J. 104, 655–665 (2013).
Erdmann, T. & Schwarz, U. S. Stochastic force generation by small ensembles of myosin II motors. Phys. Rev. Lett. 108, 188101 (2012).
Saitoh, T. et al. Differential localization of non-muscle myosin II isoforms and phosphorylated regulatory light chains in human MRC-5 fibroblasts. FEBS Lett. 509, 365–369 (2001).
Niederman, R. & Pollard, T. D. Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J. Cell Biol. 67, 72–92 (1975).
Verkhovsky, A. B., Svitkina, T. M. & Borisy, G. G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J. Cell Biol. 131, 989–1002 (1995).
Dong, C., Chen, X. & Chen, B. Standard loading profile in matrix rigidity sensing. Biophys. J. 114, 1216–1224 (2018).
Dobbie, I. et al. Elastic bending and active tilting of myosin heads during muscle contraction. Nature 396, 383–387 (1998).
Oriola, D., Gadêlha, H., Blanch-Mercader, C. & Casademunt, J. Subharmonic oscillations of collective molecular motors. Europhys. Lett. 107, 18002 (2014).
Leduc, C., Ruhnow, F., Howard, J. & Diez, S. Detection of fractional steps in cargo movement by the collective operation of kinesin-1 motors. Proc. Natl Acad. Sci. USA 104, 10847–10852 (2007).
Parmeggiani, A., Jülicher, F., Ajdari, A. & Prost, J. Energy transduction of isothermal ratchets: generic aspects and specific examples close to and far from equilibrium. Phys. Rev. E 60, 2127–43 (1999).
Rossier, O. M. et al. Force generated by actomyosin contraction builds bridges between adhesive contacts. EMBO J. 29, 1055–1068 (2010).
Luo, W. et al. Analysis of the local organization and dynamics of cellular actin networks. J. Cell Biol. 202, 1057–1073 (2013).
Yu, M. et al. Effects of mechanical stimuli on profilin- and formin-mediated actin polymerization. Nano Lett. 18, 5239–5247 (2018).
Jégou, A., Carlier, M. F. & Romet-Lemonne, G. Formin mDia1 senses and generates mechanical forces on actin filaments. Nat. Commun. 4, 1883 (2013).
Yang, Y., Nguyen, E., Mege, R.-M., Ladoux, B. & Sheetz, M. P. Local contractions regulate E-cadherin adhesions, rigidity sensing and epithelial cell sorting. https://doi.org/10.1101/318642 (2018).
Hu, S. et al. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 19, 133–141 (2017).
Ouyang, M., Yuan, C., Muisener, R. J., Boulares, A. & Koberstein, J. T. Conversion of some siloxane polymers to silicon oxide by UV/ozone photochemical processes. Chem. Mater. 12, 1591–1596 (2000).
Efimenko, K., Wallace, W. E. & Genzer, J. Surface modification of Sylgard-184 poly(dimethyl siloxane) networks by ultraviolet and ultraviolet/ozone treatment. J. Colloid Interface Sci. 254, 306–315 (2002).
Schnyder, B. et al. UV-irradiation induced modification of PDMS films investigated by XPS and spectroscopic ellipsometry. Surf. Sci. 532–535, 1067–1071 (2003).
Berdichevsky, Y., Khandurina, J., Guttman, A. & Lo, Y. H. UV/ozone modification of poly(dimethylsiloxane) microfluidic channels. Sens. Actuat. B 97, 402–408 (2004).
Song, J., Tranchida, D. & Vancso, G. J. Contact mechanics of UV/ozone-treated PDMS by AFM and JKR testing: mechanical performance from nano- to micrometer length scales. Macromolecules 41, 6757–6762 (2008).
Zheng, F. et al. The surface structure of UV exposed poly-dimethylsiloxane (PDMS) insulator studied by slow positron beam. Appl. Surf. Sci. 283, 327–331 (2013).
Schoen, I., Hu, W., Klotzsch, E. & Vogel, V. Probing cellular traction forces by micropillar arrays: contribution of substrate warping to pillar deflection. Nano Lett. 10, 1823–1830 (2010).
Li, B. et al. Development of micropost force sensor array with culture experiments for determination of cell traction forces. Cell Motil. Cytoskel. 64, 509–518 (2007).
Badoual, M., Jülicher, F. & Prost, J. Bidirectional cooperative motion of molecular motors. Proc. Natl Acad. Sci. USA 99, 6696–6701 (2002).
Guérin, T., Prost, J. & Joanny, J. F. Dynamical behavior of molecular motor assemblies in the rigid and crossbridge models. Eur. Phys. J. E 34, 60–81 (2011).
Acknowledgements
J.-F.R., J.P. and M.S. are supported by the National Research Foundation, Prime Ministers Office, Singapore and the Ministry of Education under the Research Centres of Excellence programme. In addition, J.L. and M.S. were supported by NIH grant R01GM113022.
Author information
Authors and Affiliations
Contributions
J.L., J.Hu, J.Ho. and M.P.S. designed the experimental study. J.-F.R. and J.P. designed the theoretical analysis. J.-F.R. performed the simulations. J.L. performed the experiments. J.L. and M.S. were involved in data analysis. J.Hu and J.Ho. developed the method for making dual rigidity pillars. N.M. and O.S. carried out measurements of pillar rigidity. D.P.A. implemented the 3D animations of pillar motion. J.-F.R., J.P. and M.P.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Appendices A and B, Supplementary Figures 1–10, Supplementary Tables I–III and Supplementary References 1–15.
Supplementary Video 1
Mouse embryonic fibroblast cell spreading on dual-stiffness pillars. Cyan shading represents soft pillars (3 pN nm–1, marked with C343 dye). No shading represents stiff pillars (60 pN nm–1). Pillar displacements occur primarily at the leading edge of the lamellipodium, where they can be seen as red arrows. Contractile units are highlighted by green displacement arrows. The frequency of contractile units increases as the cell enters P2 spreading. Density of contractile units per unit of cell edge, contractile unit maximum pillar displacement, and contractile unit pillar displacement velocities are invariant across stiffness lines. Movie duration 8 min (1 frame s–1).
Supplementary Video 2
Stochastic simulation of the motor assembly, with transition rates localized around the minimum of the potential. Top panel: total pillar deflection as a function of time. The red cross indicates the current state of the assembly. Bottom panel: state of the motors assembly (colour symbols) cyclic position ξi = mod(xi, l) and energy of a particular motor (blue curve) potential W1 in state 1 as a function of the cyclic coordinate ξ (orange curve) transition rate ω1 from state 1 to state 2. Parameters are K = 10 pN nm−1 and N = 120 (other parameters are given in Supplementary Table III).
Supplementary Video 3
Analytical resolution for a constant total transition rate ω of main text equation (4), corresponding to the large number of motor limit. Top panel: total pillar deflection X(t) of the collective assembly as a function of time ωt. The red cross indicates the current value of the pillar deflection. Bottom panel: joint energy–density representation — (blue curve) energy profile in state 1 as a function of the cyclic coordinate ξ = mod(x, l), (grey curve) occupation density P1 in state 1. The occupation density in state 2 is given by P2 = 1/l − P1.
Supplementary Video 4
Animation showing the process of contraction, relaxation and disassembly (performed in Blender).
Rights and permissions
About this article
Cite this article
Lohner, J., Rupprecht, JF., Hu, J. et al. Large and reversible myosin-dependent forces in rigidity sensing. Nat. Phys. 15, 689–695 (2019). https://doi.org/10.1038/s41567-019-0477-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41567-019-0477-9
This article is cited by
-
Extracellular Matrix Rigidity Modulates Human Cervical Smooth Muscle Contractility—New Insights into Premature Cervical Failure and Spontaneous Preterm Birth
Reproductive Sciences (2021)
-
Actomyosin-driven force patterning controls endocytosis at the immune synapse
Nature Communications (2019)
-
Stronger than they look
Nature Physics (2019)