Abstract
Early development of the microbiome has been shown to affect general health and physical development of the infant and, although some studies have been undertaken in high-income countries, there are few studies from low- and middle-income countries. As part of the BARNARDS study, we examined the rectal microbiota of 2,931 neonates (term used up to 60 d) with clinical signs of sepsis and of 15,217 mothers screening for blaCTX-M-15, blaNDM, blaKPC and blaOXA-48-like genes, which were detected in 56.1%, 18.5%, 0% and 4.1% of neonates’ rectal swabs and 47.1%, 4.6%, 0% and 1.6% of mothers’ rectal swabs, respectively. Carbapenemase-positive bacteria were identified by MALDI-TOF MS and showed a high diversity of bacterial species (57 distinct species/genera) which exhibited resistance to most of the antibiotics tested. Escherichia coli, Klebsiella pneumoniae and Enterobacter cloacae/E. cloacae complex, the most commonly found isolates, were subjected to whole-genome sequencing analysis and revealed close relationships between isolates from different samples, suggesting transmission of bacteria between neonates, and between neonates and mothers. Associations between the carriage of antimicrobial resistance genes (ARGs) and healthcare/environmental factors were identified, and the presence of ARGs was a predictor of neonatal sepsis and adverse birth outcomes.
Similar content being viewed by others
Main
Classically, antimicrobial resistance (AMR) is perceived as a clinical problem but non-clinical environments (for example, the human gut microbiota) are now increasingly important due to their contribution in disseminating AMR genes (ARGs). Furthermore, ARGs frequently exchange between bacteria within the human microbiota, where the intestinal bacterial community acts as a hub for horizontal gene transfer1,2. This is especially concerning for the neonatal population because colonization with multi-drug-resistant (MDR) bacteria is a precursor to invasive infections such as those leading to sepsis3,4. The incidence of neonatal sepsis and related deaths is higher in low- and middle-income countries (LMICs), which are often under-resourced to prevent, identify and treat sepsis4. Neonatal gut microbiota development and composition are shaped by the mother’s vaginal and rectal microbiotas at birth and, later, by the clinical and community environment5. The use of antibiotics, often β-lactams due to availability and cost6, perturbs the gut microbiome and can modulate bacterial populations that have a negative impact on neonatal development. Gibson et al. and other studies from Dantas’s group, primarily from high-income countries, have demonstrated that antibiotic therapy in preterm infants can dramatically affect the gut microbiome7,8,9.
Large-scale multi-national studies using molecular methods to assess the carriage of ARGs among maternal and neonatal microbiota in LMICs are non-existent. BARNARDS is a network of 12 clinical sites across 7 LMICs in Africa and south Asia aiming to assess the incidence, prevalence, risk factors, bacterial causes and burden of AMR in neonatal sepsis (https://www.ineosoxford.ox.ac.uk/research/barnards). The genomic characterization of BARNARDS’ sepsis isolates has already been discussed10, as well as their resistance profiles to β-lactam and aminoglycoside antibiotics, suggesting that the World Health Organization (WHO) may need to revise their antibiotic guidelines for neonatal sepsis within LMICs, where antibiotic resistance to current therapeutic recommendations is extremely high6.
In the present study, we characterize the Gram-negative gut microbiota of mothers and septic/non-septic neonates carrying clinically important extended-spectrum β-lactamases (ESBLs) and carbapenemase genes. We investigated statistical associations across maternal, neonatal, living environment and hospital environment domains and carriage of ESBLs and carbapenemase genes. In addition, we determined associations between neonatal/maternal carriage of ARGs, and sociodemographic and clinical environment traits. Furthermore, using whole-genome sequencing (WGS), we characterized common Gram-negative bacteria (GNBs) carrying carbapenemase genes, detailing specific variants and plasmid types across the different study sites.
Results
Prevalence of β-lactamase genes among mothers and neonates
Overall, BARNARDS recruited 35,040 mothers and their respective neonates (n = 36,285). In the present study, 18,148 rectal swabs were analysed to assess the presence of clinically important β-lactamases in the mothers’ and neonates’ gut microbiota, using the blaCTX-M-15 like gene as a marker for the presence of ESBLs and blaNDM, blaKPC and blaOXA-48-like genes, as markers of carbapenemase genes (Fig. 1).
Diagram detailing the total number of mother and neonate rectal samples collected and screened for the presence of blaCTX-M-15, blaNDM, blaKPC and blaOXA-48-like genes, the number of Gram-negative isolates carrying carbapenemase genes, the number of isolates tested for antibiotic susceptibility and the number of EC (E. coli), ENT (E. cloacae complex) and KP (K. pneumoniae) isolates characterized by WGS and bioinformatics analysis. Isolates for WGS were chosen after culture on VE (vancomycin, ertapenem) agar. Recoverable isolates after −80 °C preservation were selected for gDNA extraction and WGS.
Among 2,931 neonatal rectal swabs (BRs) analysed (our protocol aimed at collecting rectal swabs from clinically diagnosed septic neonates aged ≥7 d; however, frequently, samples were collected independently of age and all were included in the present study); 626 were from neonates with biological sepsis (BS) and 2,305 were from non-BS (NoBS) cases. The blaCTX-M-15, blaNDM, blaKPC and blaOXA-48-like genes were detected in 56.1% (within 65% of BS and 54% of NoBS), 18.5% (within 24% BS and 17% of NoBS), 0% and 4.1% (within 10% of BS and 2% of NoBS) of BRs, respectively. The prevalence of all genes was higher in south Asian countries (63.0% blaCTX-M-15, 34.7% blaNDM and 8.0% blaOXA-48-like genes) compared with African countries (49.9% blaCTX-M-15, 4.1% blaNDM and 0.6% blaOXA-48-like genes). The gene blaKPC was not detected among BRs (Fig. 2a).
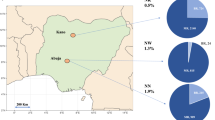
a–c, Prevalence of blaCTX-M-15, blaNDM and blaOXA-48-like genes among the rectal swabs of neonates. The prevalence of all genes was higher in South-Asian countries compared to African countries, except for blaKPC, which was not found among neonates. d–f, Prevalence of blaCTX-M-15, blaNDM and blaOXA-48-like genes among the rectal swabs of mothers. A higher prevalence of genes was seen in MR from South-Asian countries compared to African countries. blaKPC genes were found in three Indian and four Pakistani rectal samples from mothers. The BARNARDS network included the following hospitals: Bangladesh: BC and BK; Ethiopia: ES; India: IN; Nigeria: NN, NW and NK; Pakistan: PP and PC; Rwanda: RU and RK; and South Africa: ZAT. Coloured maps were created using MapChart (https://www.mapchart.net). g,h, Carriage of blaCTX-M-15, blaNDM and blaOXA-48-like genes among neonates’ rectal swabs against age of neonates at rectal swab collection per continent: Asia (g) and Africa (h). The prevalence of each ARG is plotted. The total number of samples collected per day is shown in the circles below the graphs. From day 0, ARGs were detected in the neonatal rectal microbiota. There was a tendency to a decrease in prevalence of blaNDM (53.7% to 27.7%) and blaOXA-48-like (35.4% to 0%) genes among the Asian samples through the first 14 d of life.
From 15,217 mothers’ rectal swabs (MRs) analysed, 1,299 were from mothers of neonates with BS, 13,850 from mothers of NoBS neonates and 68 from mothers with a multiple pregnancy, whose neonates had different sepsis outcomes (BSyn). From these, 47.1% (detected within 54.4% BS, 46.4% NoBS and 50% BSyn), 4.6% (detected within 6.93% BS, 4.40% NoBS and 1.47% BSyn), 0.05% (NoBS) and 1.6% (detected within 1.92% BS, 1.57% NoBS and 4.41% BSyn) carried blaCTX-M-15, blaNDM, blaKPC and blaOXA-48-like genes, respectively. A higher prevalence of genes was seen in MRs from south Asian countries (60.7% blaCTX-M-15, 8.4% blaNDM, 0.1% blaKPC and 2.4% blaOXA-48-like genes) compared with African countries (35.7% blaCTX-M-15, 1.3% blaNDM, 0% blaKPC and 1.0% blaOXA-48-like genes; Fig. 2b).
We found the prevalence of blaCTX-M-15 among MRs and BRs (Fig. 2ab) to be higher than previously reported11,12,13,14,15,16,17. The prevalence of blaOXA-48-like genes in our African sites was similar to or lower than that of other studies18,19,20. The blaOXA-48-like genes are reportedly widespread throughout south Asia21, but in our study this was observed only in Pakistan among BRs. It is interesting that blaNDM prevalence was higher than previously reported in Pakistan14,22,23, India11,24,25 and Bangladesh26. Previous reports of blaNDM neonatal carriage in Africa are few and show low-frequency rates among children and pregnant women12,20,27,28,29. Although blaKPC is widely disseminated throughout America and Europe, it is not common in south Asia or Africa11,12,18,21,24,25, as affirmed by the present study.
We analysed the neonate’s age at the time of BR collection against carriage of ARGs and found that, from day 0, ARGs were consistently found among BRs (Fig. 2c), regardless of whether delivery was via caesarean section (CS) or spontaneous vaginal delivery (SVD) and whether or not neonates developed BS (Extended Data Fig. 1a–d). A steady decrease was observed for the prevalence of blaNDM (53.7% to 27.7%) and blaOXA-48-like genes (35.4% to 0%) genes among the Asian samples through the first 14 d of life (Fig. 2c), independent of type of delivery or sepsis outcome (Extended Data Fig. 1a,c,d).
There were higher rates of ARG carriage in BS Asian neonates during the first 14 d of life (BS: blaCTX-M-15 80% (121/152), blaNDM 54% (82/152) and blaOXA-48-like genes 29% (44/152); NoBS: blaCTX-M-15 57% (250/441), blaNDM 33% (145/441) and blaOXA-48-like genes 7% (33/441)), which was also seen for African neonates, although with substantially lower differences (BS: blaCTX-M-15 58% (139/239), blaNDM 5% (12/239) and blaOXA-48-like genes 1% (3/239); NoBS: blaCTX-M-15 41% (274/674), blaNDM 3% (21/674) and blaOXA-48-like genes 1% (4/674); Extended Data Fig. 1a,b).
Similarly, among neonates born by CS in Asia, the rates of ARG carriage during the first 14 d of life were higher (CS: blaCTX-M-15 69% (178/259), blaNDM 44% (115/259) and blaOXA-48-like genes 19% (49/259); SVD: blaCTX-M-15 58% (193/334), blaNDM 34% (112/334) and blaOXA-48-like genes 8% (28/334)). This was not seen in neonates from Africa where the carriage of ARGs during the same period was similar for SVD (blaCTX-M-15 46%, 272/595; blaNDM 3%, 20/595; blaOXA-48-like genes 1%, 7/595) and CS-delivered babies (blaCTX-M-15 43%, 131/303; blaNDM 4%, 12/303; blaOXA-48-like genes 0%, 0/303; Extended Data Fig. 1c,d).
Bacterial diversity in maternal and neonatal gut microbiota
In total, 1,072 GNB isolates harbouring carbapenemase genes were recovered (Extended Data Fig. 2). From 412 BRs, we characterized 556 carbapenemase-positive bacteria (CPBs) comprising 33 species/genera with 9 isolates unidentified (Extended Data Fig. 2a). K. pneumoniae (n = 161), E. coli (n = 132) and E. cloacae complex (n = 92) were most common, accounting for 69.6% (n = 378/543) and 80.6% (n = 54/67) of positive isolates for blaNDM and blaOXA-48-like genes. K. pneumoniae and E. coli were the predominant concomitant carriers of blaNDM and blaOXA-48-like genes (n = 46/54).
Among 378 MRs, 516 CPBs from 37 distinct species/genera were characterized, with 63 isolates unidentified (Extended Data Fig. 2b). E. coli (n = 132), K. pneumoniae (n = 50) and E. cloacae complex (n = 45) were the most common, altogether accounting for 42.2% (n = 193/457) and 58.5% (n = 48/82) of positive isolates for blaNDM and blaOXA-48-like genes. K. pneumoniae and E. coli were the major carriers of blaNDM and blaOXA-48-like genes concomitantly (n = 14/21).
We found a wider CPB species diversity among BRs and MRs than previously described, where most were K. pneumoniae, E. coli and E. cloacae complex14,28,29. Evidence shows premature birth dramatically influences species richness and composition in the first months of life, enriching for E. coli, E. cloacae and Klebsiella sp.30. The 2,931 samples analysed in the present study were from 2,011 term (69%), 736 (25%) preterm and 147 (5%) post-term neonates (1% clinical data missing).
Antibiotic resistance profiles (Extended Data Fig. 3a) were established for 298 BR and 281 MR CPBs. Resistance rates were especially high for amoxicillin (97%), imipenem and ertapenem (both 91%). The gentamicin resistance rate among BR isolates was higher (84%) than for MR isolates (68%). Although high resistance rates were expected due to our selective culture method, and isolates with intrinsic resistances were recovered31, these findings contrast previous findings of low AMRs in the south Asian community32.
Genomic analysis of E. coli, E. cloacae and K. pneumoniae
The genomic epidemiology of ARGs from the dominant species among MRs and BRs (E. coli, E. cloacae complex and K. pneumoniae) was characterized by whole-genome sequencing (WGS).
From 265 E. coli, 150 isolates were sequenced (93 BRs, 57 MRs; Figs. 1 and 3) showing high genomic diversity, with 44 sequence types (STs) including one previously undefined, ST10987. Isolates were scattered across lineages when globally contextualized across E. coli from neonatal, animal, clinical and environmental samples33,34,35,36,37 (Fig. 3). Of 129 E. coli-carrying blaNDM genes, the blaNDM-5 was prominent (n = 69) across many different STs. blaOXA-181 was identified in 25 E. coli isolates from 15 STs from Bangladesh, Nigeria and Pakistan. ST405 was frequently isolated from Bangladesh (n = 24), with SNP analysis revealing distinct clades for each clinical site (Extended Data Fig. 4). Of the 13 ST405 E. coli isolates with blaNDM-5 on an IncFII plasmid (88,885 bp; Supplementary Table 1) from BK (8 BRs, 5 MRs; the sites’ acronyms are detailed in Methods), 2 isolates were from a mother–neonate pair; however, all (n = 13) were within 6 pairwise SNPs (Extended Data Fig. 4). ST405 E. coli clones have been isolated from samples from the same neonates at distinct time points, showing the persistence of this lineage in the microbiota7. A cluster of 13 BC BR ST4684 E. coli isolates were isolated within a 5-month period in 2016 (Fig. 3), all containing blaNDM-1-like genes on an IncX3 plasmid (57,221 bp; Supplementary Table 1). SNP analysis revealed this cluster to be clonal (0 pairwise SNPs). The 25 E. coli isolates carried blaOXA-181 (BC n = 2, NK n = 4, PP n = 19; 7 BRs, 18 MRs), often on a ColKP3 plasmid (n = 22; Supplementary Table 1). Notably, in ST410 from PP BRs, blaNDM-5 and blaOXA-181 were concomitantly detected, and dual-carbapenemase ARGs were also found in ST410 from global collections from the National Center for Biotechnology Information (NCBI). Of 150 BARNARDS’ CBP E. coli isolates, 90 carried blaCTX-M-15 (Fig. 3) and 4 different E. coli harboured mcr (mcr-1, n = 2 and mcr-9, n = 2).
The phylogenetic tree of 253 E. coli genomes, including 150 from BARNARDS and 103 from other studies11,12,13,14,15, is shown, using Roary (v.3.12.0)39 and FastTree40 (v.2.1.11). Isolates are coloured at the endpoint according to country and the outer ring abbreviation is labelled according to the sample source. STs for all isolates are shown in the text after the sample source. The additional two outer rings denote the presence of blaNDM and blaOXA-48-like genes. Clades containing isolates from the present study are highlighted in teal, green clades indicate E. coli neonatal sepsis isolates from other studies and pink clades relate to E. coli rectal carriage from different studies. Major STs are labelled around the phylogeny and isolates that belong to a mother–neonate pair are denoted by an orange triangle. For site acronyms, see Methods.
From 136 isolates identified by MALDI coupled to time-of-flight mass spectrometry (MALDI-TOF MS) as E. cloacae complex, 111 (77 BRs, 34 MRs) were sequenced (Figs. 1 and 4), revealing 34 STs including 5 previously undefined (Fig. 4). The E. hormaechei STs ST113, ST171 and ST418 were dominant (Fig. 4) and E. hormaechei ST418 harbouring blaNDM-1-like genes was recovered predominantly from neonates in BC and BK, with SNP analysis indicating genomic variability between 0 and 1,601 pairwise SNPs (Supplementary Table 1 and Extended Data Fig. 5). SNP analysis of 13 ST68 E. cloacae isolates from BC (8 MRs, 5 BRs), recovered during July 2016, suggests that these are very closely related (within 4 pairwise SNPs). The blaNDM-1-like gene was most common (n = 98 (BC n = 53, BK n = 17, IN n = 1, NN n = 2, PC n = 4, PP n = 21; 72 BRs, 26 MRs)) and associated with 27 different STs, often on an IncN2 or IncA/C2 plasmid (Supplementary Table 1). The blaNDM-5-like gene (n = 6) was largely identified in ST66 isolates from Bangladesh, whereas blaNDM-7 Enterobacter sp. (n = 5) was associated with four distinct STs, including previously undefined ST1372 from a Nigerian BR. blaOXA-48 variants were not detected in Enterobacter spp. In addition, 14 Enterobacter spp. isolates concomitantly carried mcr-9.1 with a variant of blaNDM-like gene (PP n = 9, PC, n = 4, BC n = 1).
The phylogenetic tree of 209 Enterobacter spp. genomes including 111 from BARNARDS and 98 from other studies12,13,15 is shown, using Roary (v.3.12.0)55 and FastTree (v.2.1.11)56. Isolates are coloured at the endpoint according to country and the outer ring abbreviation is labelled according to the sample source. STs for all isolates are shown in the text after the sample source. The additional two outer rings denote the presence of blaNDM and blaOXA-48-like genes. Clades containing isolates from the present study are highlighted in teal, green clades indicate Enterobacter spp. neonatal sepsis isolates from other studies and pink Enterobacter spp. rectal carriage from different studies. Major STs are labelled around the phylogeny. For site acronyms, see Methods.
From 211 isolates from K. pneumoniae, 161 were sequenced (125 BRs, 36 MRs; Figs. 1 and 5), including 11 K. quasipneumoniae subsp. quasipneumoniae and 9 K. quasipneumoniae subsp. similipneumoniae (Fig. 5). Three PP BR ST15 K. pneumoniae isolates possessed the same ST as that of the isolate causing sepsis in the same neonate10. We detected 46 STs from which ST11, ST14, ST15 and ST48 were common across the collective phylogeny (Fig. 5). ST11 was predominantly found in Europe carrying blaOXA-48 or blaOXA-245; however, in our study n = 6/8 PP ST11 isolates carried blaNDM-7 (Extended Data Fig. 6). The blaNDM-1 was most frequent (n = 124; Fig. 5), with 72 from Pakistan of which 4 belonged to previously undefined STs 4980–4983, and in 31 ST15 STs, the blaNDM-1 was IncA/C2 or IncN2 plasmid mediated (141,533 bp; Supplementary Table 1). In Bangladesh, 54 K. pneumoniae isolates carried blaNDM and, from these, 5 ST14 STs harboured blaNDM-1, blaOXA-232 and blaCTX-M-15. The blaOXA-232-like gene was identified on a ColKP3 plasmid in ST14 K. pneumoniae (Supplementary Table 1). Of 15 Klebsiella spp. isolates from Nigeria, 12 were K. pneumoniae, 2 K. quasipneumoniae subsp. similipneumoniae and 1 K. quasipneumoniae subsp. quasipneumoniae. Ten STs were detected, including previously undefined ST4979: nine carried blaNDM-1 and the remaining six carried blaNDM-7.
The phylogenetic tree of 268 K. pneumoniae genomes, including 161 from BARNARDS and 107 from other studies12,13,14,15,16,17,18,19,20, is shown, using Roary (v.3.12.0)55 and FastTree (v.2.1.11)56. Isolates are coloured at the endpoint according to country and the outer ring abbreviation is labelled according to the sample source. STs for all isolates are shown in the text after the sample source. The additional two outer rings denote the presence of blaNDM and blaOXA-48-like genes. Clades containing isolates from the present study are highlighted in teal, green clades indicate K. pneumoniae neonatal sepsis isolates from other studies and pink K. pneumoniae rectal carriage from different studies. Major STs are labelled around the phylogeny and isolates that belong to a mother–neonate pair are denoted by an orange triangle. Any carriage isolates sequenced in the present study that are genetically similar to the isolate recovered from the corresponding neonatal blood culture10 are denoted by a yellow star. For site acronyms, see Methods.
Risk factors for the rectal carriage of β-lactamase genes
To determine maternal, neonatal, living environment and hospital environment features associated with the carriage of blaCTX-M-15, blaNDM or blaOXA-48-like genes among the gut microbiota of mothers and neonates, we performed several exploratory univariate and multivariable analyses (Table 1 and Supplementary Table 2). We fitted a multivariable model including WASH (water, sanitation and hygiene)-associated features (Extended Data Fig. 7a), to understand the impact of these indicators in the carriage of the ARGs in the study among MRs. In 2017, 7% (Ethiopia) to 76% (South Africa) of the population in countries of the BARNARDS network used at least basic sanitation services (Supplementary Table 1) and we found that occasional handwashing by the mothers or households supplied with a wastewater network were independent risk factors for carrying blaCTX-M-15.
Multivariable models showed that occasional handwashing was associated with MR carriage of blaCTX-M-15 or blaNDM, whereas frequent handwashing was associated with the carriage of blaOXA-48-like genes (Table 1 and Supplementary Table 2a). Poor hygiene is a driver for carriage of ARGs38 and we speculate that deficient hand hygiene, even if frequent, could be associated with the carriage of these ARGs, specially blaCTX-M-15. We also found that a maternal infection in the 3 months before enrolment in the present study was associated with MR carriage of blaCTX-M-15 (Table 1 and Supplementary Table 2a). Carriage of ARGs among MRs was associated with the mothers’ use of antibiotics in the 3 months before enrolment (Table 1 and Supplementary Table 2a). In similar settings, previous use of antibiotics has been described as a risk factor for carriage of ESBL producers/MDR isolates15,38, but this was not supported from findings in other studies18,29,39. We did not find an association between neonates’ age at time of sampling and carriage of ARGs (Supplementary Table 2b). Previously, increased neonatal age was associated with carriage of ESBL producers13.
β-Lactamase gene carriage and birth outcomes
Our exploratory analysis suggests that planned or emergency CS or premature birth may be associated with the mother’s carriage of either blaCTX-M-15 or blaNDM. Also, the odds of having preterm premature rupture of membranes (PPROM) were higher for mothers carrying blaCTX-M-15 or blaNDM, whereas the odds of having perinatal asphyxia or a breech birth were higher for mothers carrying blaCTX-M-15 (Supplementary Table 2c).
As the hospital environment has been associated with carriage of ARGs19,29, we investigated whether neonates born within clinical sites (birth cohort) were more likely to carry β-lactamase genes compared with those born elsewhere (admission cohort, admitted with suspected sepsis). We found that the odds of carrying blaNDM were higher for neonates from the birth cohort (Supplementary Table 2d). We did not find significant associations between birth healthcare/environment factors and the carriage of these ARGs among the birth cohort (Extended Data Fig. 7b and Supplementary Table 2a). Univariate analysis including neonates from both cohorts showed that those born by emergency CS were more likely to carry blaCTX-M-15 in agreement with other studies13,39 and neonates born after PPROM had higher odds of carrying blaOXA-48-like genes (Supplementary Table 2e).
β-Lactamase gene carriage and neonatal sepsis
We found that colonization of the mother’s gut with blaCTX-M-15 or blaNDM positive microbiota was associated with the development of BS in the neonate, and this may be due to the mother transmitting MDR pathogens to her neonate during or after labour and birth, potentially leading to neonatal BS40. Neonates carrying blaCTX-M-15 or blaOXA-48-like genes in their microbiota were more likely to have BS compared with non-carriers (Supplementary Table 2f).
Discussion
In the present study, we report high carriage of blaCTX-M-15, blaOXA-48-like genes and blaNDM among the rectal microbiota of mothers and neonates with either suspected or confirmed BS. Carriage of genes was higher for neonates compared with mothers, as previously reported15,17, particularly, for blaNDM in samples from Bangladesh, Nigeria and Pakistan (Fig. 2). We speculate that, because most of these neonates have been administered antibiotics, if presenting with clinical sepsis, antibiotic selection pressure favoured resistant bacteria, as described previously7,15,18,19. We highlighted the carriage of ARGs in neonates from the very early hours after birth, irrespective of delivery type or sepsis outcome, which may have been underpinned by antibiotic therapy after acquisition from the mother and/or environment.
Our results further highlight the importance of access to safe water, sanitation and good hygiene to reduce the mortality rate. WASH-related factors might have been associated with the carriage of blaCTX-M-15 among MRs, and the carriage of blaCTX-M-15 or blaNDM with poor birth outcomes and neonatal sepsis. Similarly, previous maternal infection and use of antibiotics were associated with the carriage of β-lactamase genes among MRs, and further associated with more adverse birth outcomes and neonatal sepsis. In addition, our exploratory analysis suggested that complicated births such as PPROM and clinical interventions such as a CS could be associated with neonatal ARG carriage and neonatal sepsis. We acknowledge that all statistical analyses performed are exploratory and not causal. Other uncharacterized covariates such as medical history and/or socioeconomic factors are also likely to add to the AMR burden and poor health outcomes.
The genomic analysis unveiled the existence of indistinguishable E. coli isolates from MRs and BRs, suggesting transmission from mother to neonate during or after labour. Furthermore, K. pneumoniae, which was found to be the most common cause of sepsis in neonates enrolled in BARNARDS10, was also the most prevalent isolate among BRs. SNP analysis revealed three cases where K. pneumoniae BRs and sepsis isolates from the same neonate were very closely related, indicating that transmission events either in the clinical setting or in the newborn gut microbiota might have occurred. In addition, SNP analysis of E. coli, E. cloacae and K. pneumoniae genomes from neonates attending the same clinical sites indicated clonal cases. Moreover, ARGs, and in particular blaNDM-like genes, were found in different plasmids (IncX3 in Bangladesh; IncN2 or IncA/C2 in Pakistan), emphasizing a diverse dissemination of MDR pathogens harbouring ARGs. The identification of ARGs in the microbiota of neonates from the first hours of life indicates that initial colonization occurred at birth through contact with the mother and/or the hospital environment (for example, CS), and WGS analysis shows evidence for both routes of neonatal gut colonization with AMR microbiota. These findings support the need for future studies assessing mother/non-clinical environment–neonate transmission of ARGs, to improve infection prevention control measures in LMICs and study the development of the gut microbiome and resistome.
We chose β-lactamase genes as markers given the WHO recommendations of first- and second-line treatments for neonatal BS: ampicillin and ceftazidime, respectively. We acknowledge that there are many ESBL and carbapenemase genes; our selection was based on clinical importance and epidemiology. We had limitations with the retrospective recovery of E. coli, K. pneumoniae and E. cloacae for WGS due to loss of the carbapenemase gene and this may be due in part to freeze–thaw effects resulting in plasmid loss. Antibiotic susceptibility testing was performed on a proportion of recoverable isolates. One colony per phenotypically similar bacterial colony was selected for confirmation of the presence of ARGs. We acknowledge the limitation of a microbial culture-based approach that may not have detected the presence of multiple strains of the same species from a single sample. We did not perform a longitudinal study collecting samples across different time points to assess acquisition/loss of ARGs during the present study, or collect history of antibiotic exposure to understand the effects of antibiotic treatment on the neonatal microbiota. It should be noted that approximately 40% of neonatal samples discussed in the present study originate from Bangladesh (1,117/2,931, Fig. 1) and a limitation of the present study is the under-representation of available neonatal samples from other LMICs. The sociodemographic data collected and used for statistical analysis was largely self-reported and we acknowledge that this approach may have been subject to social desirability and recall bias.
In summary, the present study highlighted the prevalence of the carriage of important β-lactamase genes among the microbiota of mothers and their neonates with either suspected or confirmed sepsis in LMICs. We demonstrated the presence of ARGs in the gut microbiota from the first hours of life that has seldom been reported. We showed that poorer WASH indicators, use of antibiotics and previous infection were probably associated with gut microbiota carriage of blaCTX-M-15, blaNDM or blaOXA-48-like genes. Furthermore, the carriage of these genes was most probably associated with neonatal sepsis and adverse birth outcomes. By performing WGS on E. coli, K. pneumoniae and E. cloacae complex isolates, we unveiled the major lineages present in the guts of mothers and neonates in LMIC sites and their AMR-related genetic machinery. WGS showed relatedness between isolates from mothers’ and neonates’ microbiota and between gut microbiota and those isolates causing neonatal sepsis, warranting future studies. These results contribute to AMR surveillance in previously unexplored settings and populations to inform national action plans on better infection prevention practices and to reduce the burden of AMRs in LMICs.
Methods
Settings, ethics, participants and study design
In the present study, the term ‘neonates’ is used to include all neonates and infants (aged >28 to 60 d) enrolled. The BARNARDS network included: Bangladesh: Chittagong Maa-O-Shishu Hospital, Chattogram (BC) and Kumudini Women’s Medical College, Mirzapur (BK); Ethiopia: St Paul’s Hospital Millennium Medical College, Addis Ababa (ES); India: Division of Bacteriology, ICMR-National Institute of Cholera and Enteric Diseases Beliaghata and Institute of Post-Graduate and Medical Education & Research, Kolkata (IN); Nigeria: National Hospital Abuja (NN), Wuse District Hospital (NW) Abuja and Murtala Mohammad Specialist Hospital, Kano (NK); Pakistan: Pakistan Institute of Medical Sciences, Islamabad (PP) and Bhara Kahu Rural Health Centre, Bhara Kahu (PC); Rwanda: University Central Hospital of Kigali, Kigali (RU) and Kabgayi Hospital, Kabgayi (RK); and South Africa: Tygerberg Hospital, Cape Town (ZAT). Standard operating procedures were designed and adhered to throughout the network (https://www.ineosoxford.ox.ac.uk/research/barnards), and ethical approval was obtained from local ethics committees before the start of the study (Supplementary Table 2). The site abbreviation names were commonly used throughout this publication; however, the country name was used when the results were applicable to all sites within that country.
From November 2015 to November 2017, women in labour (preferably) or immediately post partum were recruited prospectively following their consent and their neonate(s) followed up for the first 60 d of life or until study withdrawal/neonatal death. For neonates lost to follow-up, the information available at the last follow-up point was considered. In addition, neonates who presented to clinical sites with clinically suspected sepsis in the first 60 d of life were recruited (with their mothers) on consent and followed up as described. Demographic and clinical data were collected on pretested study forms by trained researchers. The definitions for clinically suspected sepsis are detailed in https://www.ineosoxford.ox.ac.uk/research/barnards. BS was assigned to neonates with blood culture-positive sample(s), as described elsewhere10.
Further details of the study design and sociodemographic and clinical characteristics of mothers and neonates are described elsewhere41
According to the established protocol, rectal samples were to be taken from all mothers on recruitment and from neonates aged ≥7 d up to 60 d with clinically suspected sepsis. However, during the course of the present study, rectal samples were taken from neonates with clinically suspected sepsis from 0 d of life onward and these samples were also characterized and included in the present study. For this, sterile swabs in Amies Transport Medium with charcoal (Liofilchem) were used as described in https://www.ineosoxford.ox.ac.uk/research/barnards. Swabs were maintained at 4 °C until transfer to Cardiff University (CU) under UN3733 regulations at room temperature.
Ethics approval and consent to participate
Ethical approval was obtained at each of the seven participating countries (Supplementary Table 2). Bangladesh: Ethical Review Committee, Bangladesh Institute of Child Health (BICH-ERC-4/3/2015); Ethiopia: Boston Children’s Hospital (IRB-P00023058); India: Institutional Ethics Committee, National Institute of Cholera and Enteric Diseases and Institute of Post Graduate Medical Education and Research, IPGME&R Research Oversight Committee (A-I/2016-IEC and Inst/IEC/2016/508); Nigeria: Kano State Hospitals Management Board (8/10/1437AH), Health Research Ethics Committee (HREC) and National Hospital, Abuja (NHA/EC/017/2015); Pakistan: Shaheed Zulfiqar Ali Bhutto Medical University, Pakistan Institute of Medical Sciences (PIMS), Islamabad (ref. no. NA, signed letter from T. Hazir); Rwanda: Republic of Rwanda, National Ethics Committee (No342/RNEC/2015); and South Africa: Stellenbosch University and Tygerberg Hospital, Research projects, Western Cape Government (N15/07/063). All approval dates are listed in Supplementary Table 2. In local languages, research nurses provided mothers with study information and collected consent for mother and/or neonatal enrolment. Informed consent was obtained in writing unless this was not possible (due to literacy barriers) and oral consent was collected from the mothers by trained researchers. Oral consent was documented by the participant signing/marking the consent form.
Gut microbiota characterization
On arrival to CU, rectal swabs were stored at 4 °C until processing. Mothers’ rectal samples were processed on a ratio of a minimum of 1:3 BS:NoBS-related sample per site10 and all neonatal rectal swabs were processed. Swabs were streaked on three chromogenic agar medium plates (Liofilchem) supplemented with either vancomycin (10 mg l−1), vancomycin and cefotaxime (VC, 10 mg l−1 and 1 mg l−1, respectively) and vancomycin and ertapenem (VE, 10 mg l−1 and 2 mg l−1, respectively) to select for cefotaxime-resistant GNBs (indicative of the presence of ESBL producers) and ertapenem-resistant GNBs (indicative of the presence of carbapenemase producers).
The GNB microbiota grown on VC and VE plates was scrutinized for the presence of blaCTX-M-15 and of blaNDM, blaKPC and blaOXA-48-like genes, correspondingly, by PCR/multiplex-PCR using the Illustra PuReTaq Ready-To-Go PCR Beads (GE Healthcare) in a Gene Touch Thermal Cycler (Hangzhou Bioer Technology Co., Ltd). PCR conditions, primers (Eurofins) and control strains are described in Supplementary Table 3. Amplicons were subjected to electrophoresis in a 1% agarose (Sigma-Aldrich) gel at 300 V for 35 min in 1× Tris/borate/EDTA buffer containing 25 µl of ethidium bromide. All bacterial cultures were preserved in TS/72 beads (Technical Service Consultants) at −80 °C.
Phenotypically distinct bacterial colonies in VE plates from multiplex-PCR-positive samples were selected and pure cultures obtained by repeated isolation of individual colonies in the same medium. All isolates were subjected to multiplex-PCR and those with a positive result for any of the carbapenemase genes in the study were identified by MALDI-TOF MS (Bruker Daltonik GmbH) and preserved as mentioned before until further analysis. The workflow for sample collection and processing is shown in Extended Data Fig. 8. Due to the high prevalence of blaCTX-M-15, we did not scrutinize samples for blaCTX-M-15-positive isolates.
Indian samples were processed locally using the same methodology, except for bacterial isolate identification, which was done using Enterosystem 18R (Liofilchem) and the VITEK 2 Compact Automated System.
BSyn sample results were included in both BS and NoBS groups, because the same mother had neonates with different BS statuses. Hence, the results for each of the 68 samples were accounted for twice.
AMR profiles
Antibiotic susceptibility testing was performed using the disk diffusion method for a subset of isolates (n = 584) according to EUCAST v.9 guidelines (2019)31, using appropriate control strains to test quality control. Antibiotics tested were tigecycline (TGC, 15 μg), fosfomycin (FOS, 200 μg), ciprofloxacin (CIP, 5 μg), levofloxacin (LVX, 5 μg), gentamicin (GEN, 10 μg), amikacin (AMK, 30 μg), nitrofurantoin (F, 100 μg), trimethoprim–sulfamethoxazole (SxT, 1.25/23.75 μg), ertapenem (ETP, 10 μg), amoxicillin (AML, 10 μg), amoxicillin–clavulanic acid (AMC, 20/10 μg), piperacillin–tazobactam (TZP, 30/6 μg), CTX (cefotaxime, 5 μg), ceftazidime (CAZ, 10 μg), cefepime (FEP, 30 μg), imipenem (IPM, 10 μg), meropenem (MEM, 10 μg) and aztreonam (ATM, 30 μg). Indian bacterial isolates were tested locally using the same methods. Supplementary Table 4 shows the antibiotics tested and the disk concentrations, control strains used and EUCAST v.9 breakpoint tables used for interpretation of results.
Genomic analysis of E. coli, E. cloacae and K. pneumoniae
K. pneumoniae, E. coli and E. cloacae isolates from rectal swabs from Bangladesh, Pakistan and Nigeria were selected for further characterization by WGS and bioinformatics analysis. Genomic (g)DNA extraction and Illumina WGS were performed as described10.
Briefly, gDNA was extracted using the QIAmp DNA mini-kit (QIAGEN), with an additional RNase step, on the automated QIAcube platform (QIAGEN), and was quantified using the Qubit fluorometer 3.0. Genomic libraries were prepared using Nextera XT v.2 (Illumina), with a bead-based normalization, following the manufacturers’ guidelines. A total of 48 isolates is multiplexed per sequencing run to provide a depth of coverage >15×. Paired-end WGS was performed on an Illumina MiSeq using the v.3 chemistry to generate fragment lengths up to 300 bp (600 cycles). For Oxford Nanopore Technology (ONT) sequencing, fresh gDNA was extracted as described above, concentrated using SPRI beads (Mag-Bind TotalPure, Omega) and libraries were generated using the 96-Rapid Barcoding Kit (SQK-RBK110.96; ONT). Sequencing was performed using MinION flow cells (R9.4 and R.10) for a running time of 72 h within MinKnow.
Bioinformatics analysis was performed using a high-performance computing cluster at CU (Advanced Research Computing at Cardiff (ARCCA)) and CLIMB42. Paired-end reads (fastq) were subjected to quality control checks before downstream analysis. Trimgalore (v.0.4.3)43 was used to remove the Nextera adaptor sequences and low-quality bases. Reports before and after read trimming were generated using fastqc (v.0.11.2)44 and collated using MultiQC (v.1.7)45. The mean read length and number of sequences provided on the MultiQC reports were used to determine sequencing coverage. Paired-end reads were assembled using the Shovill pipeline with associated dependencies. Final genome assembly metrics were generated using QUAST (v.2.1)46. Bacterial species were identified using BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (v.2.2.25)47 (input: contigs) and PathogenWatch48. Multilocus sequence typing (MLST), antibiotic resistance and plasmid genomic profiles were characterized using ABRicate v.0.9.7 (ref. 49) and associated databases: NCBI50 and PlasmidFinder51. The average nucleotide identity (ANI) was calculated using ChunLab’s online ANI52.
Previously undefined alleles and ST profiles were submitted to Enterobase, BIGSbd and PubMLST for assignment53. Genomes were annotated using Prokka (v.1.12)54. Isolate relatedness analysis was performed using Roary (v.3.12.0)55 to create a core genome alignment and FastTree (v.3.12.0)56 to generate maximum likelihood phylogenetic trees. Core phylogenetic trees were mid-rooted, visualized and annotated using iTOL (v.4)57.
SNP analysis was performed on ST-specific clades using snippy (v.4.6.0)58 (input paired-end fastq) with BWA and freebayes mapping the reads and calling variants. To maximize SNP calling, for each clade, a high-quality reference was generated59 using long reads (ONT bioinformatics, see below; Supplementary Table 5 summarizes the genome metrics for each SNP reference genome). Snippy-core was used to concatenate SNPs and snp-sites60 was used to extract SNPs. Gubbins (v.2.3.4)61 was used to identify and remove recombination. IQ-tree (v.2.0) was used to generate a maximum likelihood SNP tree62. Snp-dists was used to generate a pairwise SNP matrix63. SNP trees were outgroup rooted where possible, or mid-point rooted and visualized with iTOL (v.4)57.
ONT FAST5 reads were base called using Guppy v.5.0.11 and NVIDIA v.100 GPUs. Filtlong (v.0.2.0) was used to trim fastq (--min_length 1000 --keep_percent 90) and the reads were assembled with the corresponding short reads using Unicycler v.0.4.9 with default parameters. The number and length (N50) of long reads was determined using Nanoplot (v.1.19.0)64. Plasmid sequences were extracted using Bandage v.0.8.11 (ref. 65) and assessed for similarity using PLSDB66 and BLAST47. Plasmid analysis was performed for n = 50 isolates chosen based on short-read sequencing analysis (ARG carriage, ST and core genome phylogeny) and other metadata available (clinical site, sample type, date).
Illumina paired-end sequence reads were submitted to the European Nucleotide Archive (ENA) and given accession no. PRJEB39293. Hybrid genomes (Illumina and ONT) were submitted to the NCBI and given BioProject accession no. PRJNA767644.
Global isolates for contextual analysis
Approximately 100 isolates of E. coli, K. pneumoniae, and E. cloacae were included in phylogenetic analyses. Isolates were chosen from two searches. First, a literature search ascertained the availability of whole genomes from studies focusing on neonatal studies and/or rectal or intestinal carriage of ESBL/carbapenemase, primarily, but not exclusively in LMICs. Available clinical and other associated data, including country, sample, source and date, were collected, where available.
Second, to provide further context from additional countries and sources, including animal and environmental, between 50 and 80 genomes were chosen from the NCBI Assembly collection. On 5 March 2020 sequence data in fasta file format were downloaded from the NCBI’s Assembly resource. For E. coli, 18,761 genomes were downloaded and 18,673 were further identified as E. coli using in-house bioinformatics analysis as described above. A total of 1,790 different STs were found within the E. coli collection. For K. pneumoniae, 8,663 genomes were downloaded and 8,660 were further identified as K. pneumoniae using in-house bioinformatics analysis. A total of 930 different STs were found within the K. pneumoniae collection. For E. cloacae complex, 1,960 genomes were downloaded and 1,886 were further identified as being E. cloacae complex, of which 398 different STs were found.
ABRicate v.0.9.7 was used to screen all genomes for ARGs. To assist choosing both blaNDM/OXA-48-like-positive and -negative isolates, the dataset was divided according to the ARG output. Genomes were then chosen at random and the accession nos. used to obtain biosample information, including, where possible, country, sample, source and date of isolate.
Statistical analysis
A formal sample size was not calculated. Sites were asked to recruit all eligible mothers into the overarching study over a period of at least 12 months. All BRs were processed and MRs were processed at a ratio previously described.
Logistic regression models were fitted to maternal and living environment variables (maternal carriage) and healthcare settings, maternal and living environment variables (neonate carriage) to investigate associations with MR and BR β-lactamase gene carriage (blaCTX-M-15, blaNDM and blaOXA-48-like genes separately). These were also done for the neonate birth cohort only. Several multivariable analyses were also carried out to explore the association between these variables and maternal/infant carriage of ARGs. Explanatory variables were selected for inclusion in multivariable models on the basis of expert opinion and literature67. The variables included are detailed in Supplementary Methods.
We investigated the association between MR/BR β-lactamase gene carriage and neonatal BS by fitting logistic regression models.
To investigate the association between MR β-lactamase gene carriage and birth outcomes (delivery type, timing of birth, perinatal asphyxia, breech presentation and PPROM), multinomial (delivery type (SVD as the base outcome), timing of birth (on time as the base outcome), perinatal asphyxia (no as the base outcome)) and logistic regression (breech presentation and PPROM) models were fitted with birth outcomes as the outcome, and MR β-lactamase gene carriage as the explanatory variable. Conversely, to investigate the association between birth outcomes and BR β-lactamase gene carriage, logistic regression models were fitted with BR β-lactamase gene carriage as the outcome and birth outcomes as the explanatory variable.
All models were adjusted for site as a fixed effect. For the association between β-lactamase gene carriage and neonatal BS, we also reported the associations without adjusting for site. Logistic regression models are reported as odds ratios (ORs), 95% confidence intervals (CIs) and P values. Multinomial logistic regression models are reported as relative risk ratios, 95% CIs and P values. P values were adjusted for multiple testing using the Holm–Bonferroni method68 on a per outcome/model basis (for example, associations with blaCTX-M-15 in MRs were adjusted for separately to associations with blaNDM in MRs, and so on), with a familywise error rate (FWER) of 0.05. Furthermore, owing to the small percentage of missing data, no imputation of missing variables was performed. Given the large number of hypothesis tests reported in the present study, findings where Padj < 0.05 are highlighted in the main text. However, findings from all analyses can be found in Table 1 and the accompanying data, and Supplementary Methods. All analyses used the z-test from a logistic regression model and all statistical tests were two sided. Statistical analyses were conducted using Stata v.16.1.
Extended Data Fig. 7 was edited to reflect significant values with coloured dots using Adobe Illustrator v.25.0.1.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequence reads have been submitted to the European Nucleotide Archive under accession no. PRJEB39293. Individual accession nos. and additional genomics data can be accessed in the Supplementary Methods and in the Source data provided with this paper. Hybrid assemblies (Illumina and ONT) have been submitted to the NCBI under the BioProject accession no. PRJNA767644. Databases used within this study: VFDB, http://www.mgc.ac.cn/VFs/download.htm; NCBI, https://github.com/tseemann/abricate/tree/master/db/ncbi; Resfinder, https://github.com/tseemann/abricate/tree/master/db/resfinder; Plasmidfinder, https://bitbucket.org/genomicepidemiology/plasmidfinder/src/master; mlst, https://github.com/tseemann/mlst/tree/master/db/pubmlst; PLSBD, https://ccb-microbe.cs.uni-saarland.de/plsdb.
Code availability
Programs were used with default parameters unless specified. The code used is available upon request.
References
Trobos, M., Lester, C. H., Olsen, J. E., Frimodt-Moller, N. & Hammerum, A. M. Natural transfer of sulphonamide and ampicillin resistance between Escherichia coli residing in the human intestine. J. Antimicrob. Chemother. 63, 80–86 (2008).
Porse, A. et al. Genome dynamics of Escherichia coli during antibiotic treatment: transfer, loss, and persistence of genetic elements in situ of the infant gut. Front. Cell. Infect. Microbiol. https://doi.org/10.3389/fcimb.2017.00126 (2017).
Shane, A. L., Sánchez, P. J. & Stoll, B. J. Neonatal sepsis. Lancet 390, 1770–1780 (2017).
Rudd, K. E. et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 395, 200–211 (2020).
Shao, Y. et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121 (2019).
Thomson, K. M. et al. Effects of antibiotic resistance, drug target attainment, bacterial pathogenicity and virulence, and antibiotic access and affordability on outcomes in neonatal sepsis: an international microbiology and drug evaluation prospective substudy (BARNARDS). Lancet Infect. Dis. 21, 1677–1688 (2021).
Gasparrini, A. J. et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat. Microbiol. 4, 2285–2297 (2019).
Gibson, M. K. et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat. Microbiol. 1, 16024 (2016).
Gasparrini, A. J. et al. Antibiotic perturbation of the preterm infant gut microbiome and resistome. Gut Microbes 7, 443–449 (2016).
Sands, K. et al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat. Microbiol. 6, 512–523 (2021).
Kothari, C. et al. Community acquisition of β-lactamase producing Enterobacteriaceae in neonatal gut. BMC Microbiol. 13, 1 (2013).
Berendes, D. et al. Gut carriage of antimicrobial resistance genes among young children in urban Maputo, Mozambique: associations with enteric pathogen carriage and environmental risk factors. PLoS ONE 14, e0225464 (2019).
Kagia, N. et al. Carriage and acquisition of extended-spectrum β-lactamase-producing Enterobacterales among neonates admitted to hospital in Kilifi, Kenya. Clin. Infect. Dis. 69, 751–759 (2019).
Saleem, A. F. et al. The gut of healthy infants in the community as a reservoir of esbl and carbapenemase-producing bacteria. Antibiotics 9, 1–11 (2020).
Kurz, M. S. E. et al. Intense pre-admission carriage and further acquisition of ESBL-producing Enterobacteriaceae among patients and their caregivers in a tertiary hospital in Rwanda. Trop. Med. Int. Health 22, 210–220 (2017).
Farra, A. et al. High rate of faecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae in healthy children in Bangui, Central African Republic. Clin. Microbiol. Infect. 22, 891.e1–891.e4 (2016).
Desta, K. et al. High gastrointestinal colonization rate with extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS ONE 11, 1–14 (2016).
Labi, A.-K. et al. High Carriage rates of multidrug-resistant Gram-negative bacteria in neonatal intensive care units from Ghana. Open Forum Infect. Dis. 7, ofaa109 (2020).
Mairi, A. et al. Carbapenemase-producing Enterobacteriaceae among pregnant women and newborns in Algeria: prevalence, molecular characterization, maternal-neonatal transmission, and risk factors for carriage. Am. J. Infect. Control 47, 105–108 (2019).
Kieffer, N., Nordmann, P., Aires-De-Sousa, M. & Poirel, L. High prevalence of carbapenemase-producing Enterobacteriaceae among hospitalized children in Luanda, Angola. Antimicrob. Agents Chemother. 60, 6189–6192 (2016).
van Duin, D. & Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8, 460–469 (2017).
Day, K. M. et al. Prevalence and molecular characterization of Enterobacteriaceae producing NDM-1 carbapenemase at a military hospital in Pakistan and evaluation of two chromogenic media. Diagn. Microbiol. and Infect. Dis. 75, 187–191 (2013).
Perry, J. D. et al. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66, 2288–2294 (2011).
Mittal, G. et al. Risk factors for fecal carriage of carbapenemase producing Enterobacteriaceae among intensive care unit patients from a tertiary care center in India. BMC Microbiol. 16, 1–10 (2016).
Antony, S., Ravichandran, K. & Kanungo, R. Multidrug-resistant Enterobacteriaceae colonising the gut of adult rural population in South India. Indian J. Med. Microbiol. 36, 488–93 (2018).
Islam, M. A. et al. Prevalence of faecal carriage of NDM-1-producing bacteria among patients with diarrhoea in Bangladesh. J. Med. Microbiol. 63, 620–622 (2014).
Ramsamy, Y. et al. Genomic analysis of carbapenemase-producing extensively drug-resistant Klebsiella pneumonia isolates reveals the horizontal spread of p18-43_01 plasmid encoding blandm-1 in South Africa. Microorganisms 8, 1–16 (2020).
Ogbolu, D. O. & Webber, M. A. High-level and novel mechanisms of carbapenem resistance in Gram-negative bacteria from tertiary hospitals in Nigeria. Int. J. Antimicrob. Agents 43, 412–417 (2014).
Chereau, F. et al. Colonization of extended-spectrum-β-lactamase- and NDM-1-producing Enterobacteriaceae among pregnant women in the community in a low-income country: a potential reservoir for transmission of multiresistant Enterobacteriaceae to neonates. Antimicrob. Agents Chemother. 59, 3652–3655 (2015).
Henderickx, J. G. E., Zwittink, R. D., van Lingen, R. A., Knol, J. & Belzer, C. The preterm gut microbiota: an inconspicuous challenge in nutritional neonatal care. Front. Cell. Infect. Microbiol. https://doi.org/10.3389/fcimb.2019.00085 (2019).
European Committee on Antimicrobial Susceptibility Testing. EUCAST v.9. Breakpoint tables for interpretation of MICs and zone diameters. v.9.0 (2019). http://www.eucast.org
Chaurasia, S. et al. Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ 364, k5314 (2019).
Irenge, L. M. et al. Whole-genome sequences of multidrug-resistant Escherichia coli in South-Kivu Province, Democratic Republic of Congo: characterization of phylogenomic changes, virulence and resistance genes. BMC Infect. Dis. 19, 1–10 (2019).
Sugawara, Y. et al. Spreading patterns of NDM-producing Enterobacteriaceae in clinical and environmental settings in Yangon, Myanmar. Antimicrob. Agents Chemother. 63, e01924-18 (2019).
Stoesser, N. et al. Genome sequencing of an extended series of NDM-producing Klebsiella pneumoniae isolates from neonatal infections in a Nepali hospital characterizes the extent of community- versus hospital-associated transmission in an endemic setting. Antimicrob. Agents Chemother. 58, 7347–7357 (2014).
Runcharoen, C. et al. Whole genome sequencing reveals high-resolution epidemiological links between clinical and environmental Klebsiella pneumoniae. Genome Med. 9, 1–10 (2017).
Founou, L. L. et al. Genome sequencing of extended-spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae isolated from pigs and abattoir workers in Cameroon. Front. Microbiol. 9, 1–12 (2018).
Pathak, A., Chandran, S. P., Mahadik, K., Macaden, R. & Lundborg, C. S. Frequency and factors associated with carriage of multi-drug resistant commensal Escherichia coli among women attending antenatal clinics in Central India. BMC Infect. Dis. 13, 199 (2013).
Herindrainy, P. et al. Rectal carriage of extended-spectrum beta-lactamase-producing Gram-negative bacilli in community settings in Madagascar. PLoS ONE https://doi.org/10.1371/journal.pone.0022738 (2011).
Chan, G. J., Lee, A. C., Baqui, A. H., Tan, J. & Black, R. E. Risk of early-onset neonatal infection with maternal infection or colonization: a global systematic review and meta-analysis. PLoS Med. 10, e1001502 (2013).
Milton, R. et al. Neonatal sepsis and mortality in low-income and middle-income countries from a facility-based birth cohort: an international multisite prospective observational study. Lancet Global Health 10, e661–e672 (2022).
Connor, T. R. et al. CLIMB (the Cloud Infrastructure for Microbial Bioinformatics): an online resource for the medical microbiology community. Microb. Genom. 2, e000086 (2016).
Krueger, F. Trim Galore.
Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
Ewels, P., Magnusson, M., Lundin, S. & Käller, M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016).
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075 (2013).
Mcginnis, S. & Madden, T. L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 32, 20–25 (2004).
Pathogenwatch A Global Platform for Genomic Surveillance developed by Centre for Genomic Pathogen Surveillance. https://pathogen.watch/
Seemann, T. ABRicate. https://github.com/tseemann/abricate
Feldgarden, M. et al. Validating the AMRFINder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 63, 1–19 (2019).
Carattoli, A. et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Yoon, S.-H. H., Ha, S. M., Lim, J., Kwon, S. & Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286 (2017).
Maiden, M. C. J. et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat. Rev. Microbiol. 11, 728–736 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Page, A. J. et al. Sequence analysis Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693 (2015).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, 256–259 (2019).
Seemann, T. snippy: fast bacterial variant calling from NGS reads (2015). https://github.com/tseemann/snippy
Bush, S. J. et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. Gigascience 9, 1–21 (2020).
Page, A. J. et al. SNP-sites: rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2, e000056 (2016).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015).
Minh, B. Q. et al. IQ-TREE 2: new models and efficient methods for phylogenetic iInference in the genomic era. Mol. Biol. Evol. 37, 1530–1534 (2020).
Seemann, T. snp-dists: Pairwise SNP distance matrix from a FASTA sequence alignment. https://github.com/tseemann/snp-dists
de Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018).
Wick, R. R., Schultz, M. B., Zobel, J. & Holt, K. E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352 (2015).
Galata, V., Fehlmann, T., Backes, C. & Keller, A. PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202 (2019).
Wallisch, C. et al. Review of guidance papers on regression modeling in statistical series of medical journals. PLoS ONE 17, e0262918 (2022).
Holm, S.A simple sequentially rejective multiple test procedure. Scand. J. Statist. 6, 65–70 (1979).
Acknowledgements
We thank all participants and their families. This work was supported by a combination of two research awards (nos. OPP1119772 and OP1191522) from the Bill & Melinda Gates Foundation. We thank Liofilchem for their continued support in the distribution of their microbiology products to enable standardization of standard operating procedures across the clinical sites. We thank J. Parkhill for advice and guidance regarding the phylogenetic analyses. We thank Wales Gene Park and ARCCA for their continued bioinformatics support and infrastructure availability. Bioinformatics analysis was largely undertaken using the supercomputing facilities at CU operated by ARCCA on behalf of the Cardiff Supercomputing Facility and the HPC Wales and Supercomputing Wales projects. The latter is part funded by the European Regional Development Fund via the Welsh Government. We thank the team of curators for the databases hosted on PubMLST https://pubmlst.org/databases. We thank the curators of the Institut Pasteur MLST and whole-genome MLST databases for curating the Klebsiella spp. data and making them publicly available at http://bigsdb.pasteur.fr. We thank M. Islam for providing access to the clinical sites and epidemiology data in Bangladesh. We would like to acknowledge R. Kamran, the microbiologist from PIMS, Pakistan, who sadly passed away in 2018. We thank the team within the Specialist Antimicrobial Chemotherapy Unit, University Hospital Wales, Public Health Wales for their support for MALDI-TOF MS of bacterial isolates. We thank A. Reis (iBiMED, University of Aveior) for help in producing Fig. 2c and Extended Data Fig. 1.
Author information
Authors and Affiliations
Consortia
Contributions
M.J.C. and K.S. designed, guided the study and analysis, and wrote the manuscript. K.S., E.P. and I.B. performed the WGS experiments. M.J.C., K.S., K. Thomson, E.P., J.M., C.D., C.A., P.H., H.S., A.F., M.N., T.H., S.N., A.M., A.R. and L.R. performed the microbiology experiments. M.J.C. and B.H. optimized the PCR reaction. K.S., M.J.C. and R.A. performed the bioinformatics analysis. R.M., D.G., K.H., G.C., C.D. and T.R.W. designed and delivered the epidemiological aspects of the study. J.W. performed analysis to guide sample selection and processing. D.G. performed statistical analysis. G.C., D.B., S.S., G.M., S.B., P.C., S.H., A.S., S.M., K.I., F.M., S.U., L.A., C.E., A.H.Y., A.A., A.S.M., R.Z., H.S., A.M., N.S., M.H.J., S.A., J.B.M., A.R., L.G., S.M., A.N.H.B., A.W. and L.M. assisted in collecting rectal swab samples and transporting to the United Kingdom. G.C., S.B., K.I., R.Z., J.B.M. and S.M. facilitated the epidemiology data collection at the clinical sites. T.R.W., M.J.C., R.M., G.C., S.B., K.I., R.Z., J.B.M. and S.M. designed the BARNARDS study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks Amy Mathers and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Carriage of blaCTX-M-15, blaNDM and blaOXA-48-like genes among neonates’ rectal swabs by age of neonates at sample collection.
Carriage of blaCTX-M-15, blaNDM and blaOXA-48-like genes among neonates’ rectal swabs against age of neonates at sample collection per biological sepsis status in a) Asia and b) Africa and per delivery type in c) Asia and d) Africa. Prevalences of each antibiotic resistance gene (ARG) are plotted. The total number of samples collected per day for each type of delivery is shown in the circles below the graphs. From day 0, ARGs were detected in the neonates’ faecal microbiota independently of biological sepsis status or delivery type. Plots were done in R studio using packages tidyr (v1.2.0), ggpubr (v0.4.3), gridExtra (v2.3), and egg (v0.4.3).
Extended Data Fig. 2 Species diversity of the isolates recovered from rectal samples.
Diversity of carbapenemase positive Gram-negative bacterial isolates collected from a) neonates’ and b) mothers’ rectal swabs. The number of isolates collected per species is shown.
Extended Data Fig. 3 Antimicrobial resistance profiles of the isolates recovered from rectal samples.
Antimicrobial resistance (AMR) profiles of the carbapenemase positive Gram-negative bacterial isolates collected from a) neonates’ and b) mothers’ rectal swabs per species. AMR profiles distributed by site for c) neonates’ and d) mothers’ isolates distributed by site are also shown. The number of isolates tested for antibiotic susceptibility per species (a, b) and per site (c, d) is shown in the legends. The overall percentage of resistant isolates to each antimicrobial tested is shown at the top of the corresponding bar. For antibiotics’ acronyms see methods.
Extended Data Fig. 4 Whole genome single-nucleotide polymorphisms analysis of ST405 E. coli isolates.
Whole genome SNP analysis of ST405 E. coli (n = 24 isolates) using snippy v4.6.0 on paired end fastq, gubbins v2.3.4 to remove recombination and IQ-tree v2.0 to construct the phylogeny. iTOL v4 was used to visualise the phylogenetic tree. BCBR/BCMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Chittagong Maa-O-Shishu Hospital, Chattogram (BC). BKBR/BKMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Kumudini Women’s Medical College, Mirzapur (BK). PPBR, neonate (BR) rectal isolates from Pakistan Institute of Medical Sciences (PP), Islamabad. Reference, BC-MR1421-3. Dates of rectal swab collection are shown.
Extended Data Fig. 5 Whole genome single-nucleotide polymorphisms analysis of ST11 and ST15 K. pneumoniae isolates.
a) SNP analysis of ST11 Klebsiella pneumoniae (n = 12 isolates) collected during this study and K. pneumoniae GCF_001462965, GCF_006351115 and GCF_900607955 (see SourceData for Enterobacter sp. trees) using snippy v4.6.0 on paired end fastq, gubbins v2.3.4 to remove recombination and IQ-tree v2.0 to construct the phylogeny. iTOL v4 was used to visualise the phylogenetic tree. BCBR/BCMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Chittagong Maa-O-Shishu Hospital, Chattogram (BC). BKBR/BKMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Kumudini Women’s Medical College, Mirzapur (BK). PPBR/PPMR, neonate (BR)/mother (MR) rectal isolates from Pakistan Institute of Medical Sciences (PP), Islamabad. Reference, PP-BR254-2. Dates of rectal swab collection are shown. b) SNP analysis of ST15 K. pneumoniae (n = 25 isolates; BARN) collected during this study and other K. pneumoniae isolates’ genome assemblies available in GeneBank (see SourceData for Klebsiella pneumoniae trees) using snippy v4.6.0 on paired end fastq, gubbins v2.3.4 to remove recombination and IQ-tree v2.0 to construct the phylogeny. iTOL v4 was used to visualise the phylogenetic tree. PPBR/PPMR, neonate (BR)/mother (MR) rectal isolates from Pakistan Institute of Medical Sciences (PP), Islamabad. Reference, PP-BR737-1. Dates of rectal swab collection are shown.
Extended Data Fig. 6 Whole genome single-nucleotide polymorphisms analysis of ST418 E. hormaechei isolates.
SNP analysis of ST418 E. hormaechei (n = 16) collected during this study and Enterobacter hormaechei subsp. xiangfangensis GCF_1526085 using snippy v4.6.0 on paired end fastq, gubbins v2.3.4 to remove recombination and IQ-tree v2.0 to construct the phylogeny. iTOL v4 was used to visualise the phylogenetic tree. BCBR/BCMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Chittagong Maa-O-Shishu Hospital, Chattogram (BC). BKBR/BKMR, neonate (BR)/mother (MR) rectal isolates from Bangladesh, Kumudini Women’s Medical College, Mirzapur (BK). Reference, BC-MR78-3. Dates of rectal swab collection are shown.
Extended Data Fig. 7 Exploratory multivariable statistical analysis to identify associations between socio-demographic and clinical data and maternal and neonatal carriage of ARGs.
Forest plots representing exploratory multivariable statistical analysis to identify associations between socio-demographic and clinical data and maternal and neonatal carriage of ARGs. Bars represent ranges of odds ratio. Multiplicity-adjusted p-values that remained statistically significant at the 5% level are coloured in red. Z tests were used from multivariable logistic regression models and statistical tests were two-sided. a) Two multivariable models (MV) were performed to understand the association between WASH (water, sanitation and hygiene) related variables and maternal carriage of ARGs using the explanatory variables shown. MV2 results are displayed; In MV1, type of toilet in home did not gave an association with carriage of any of the ARGs. b) Two MV were performed to understand the association between birth healthcare environment features and carriage of ARGs among neonates from the birth cohort using the explanatory variables shown. MV1 results are shown. Perinatal asphyxia is not plotted given the wide confidence intervals.
Extended Data Fig. 8 BARNARDS workflow for collection of samples and data in local sites and samples and isolates characterisation in Cardiff University.
Indian samples characterisation was performed locally.
Supplementary information
Supplementary Information
Supplementary Methods and Tables 1–5.
Supplementary Table 1
Data on plasmid types harbouring blaNDM and blaOXA-48-like genes unveiled by hybrid Illumina and ONT genome assembly.
Supplementary Table 2
Workbook with multiple tabs (a–f) containing results of statistical analyses performed. Legends are available in each tab.
Source data
Source Data Fig. 1
Number of mothers and neonates enrolled, MRs and BRs collected and analysed, positive samples for the β-lactamase genes in study and number of isolates recovered and positive for the carbapenemase genes in study per site.
Source Data Fig. 2
a,b, Number of mothers and neonates enrolled, MRs and BRs collected and analysed, positive samples for the β-lactamase genes in study and number of isolates recovered and positive for the carbapenemase genes in study per site and BS status. c, Number of samples positive for the β-lactamase genes in study, per continent (Asia and Africa), distributed by infants’ age at rectal sample collection. The total number of samples collected per day distributed by infants’ age is also shown.
Source Data Fig. 3
E. coli phylogenetic tree.
Source Data Fig. 4
K. pneumoniae phylogenetic tree.
Source Data Fig. 5
Enterobacter spp. phylogenetic tree.
Source Data Extended Data Fig. 1
a,b, Number of samples positive for the β-lactamase genes in study, per continent (Asia and Africa) and BS status, distributed by infants’ age at rectal sample collection. The total number of samples collected per day distributed by infants’ age is also shown. c,d, Number of samples positive for the β-lactamase genes in study, per continent (Asia and Africa) and type of delivery, distributed by infants’ age at rectal sample collection. The total number of samples collected per day distributed by infants’ age is also shown.
Source Data Extended Data Fig. 2
Genus/species identification of bacterial isolates and respective overall numbers recovered from rectal samples (neonates and mothers) positive for the carbapenemase genes in study per site and according to BS status. Isolates were identified by MALDI-TOF MS, Enterosystem 18R (Liofilchem) or VITEK 2 Compact Automated System. a, Genus/species identification of bacterial isolates and respective numbers recovered from BRs positive for the carbapenemase genes in study per site and according to BS statuts. Isolates were identified by MALDI-TOF MS, Enterosystem 18R (Liofilchem) or VITEK 2 Compact Automated System. b, Genus/species identification of bacterial isolates and respective numbers recovered from MRs positive for the carbapenemase genes in study per site and according to BS status. Isolates were identified by MALDI-TOF MS, Enterosystem 18R (Liofilchem) or VITEK 2 Compact Automated System.
Source Data Extended Data Fig. 3
a, Number of antibiotic-resistant isolates collected from BRs per species. b, Number of antibiotic-resistant isolates collected from MRs per species. c, Number of antibiotic-resistant isolates collected from BRs per site. d, Number of antibiotic-resistant isolates collected from MRs per site.
Source Data Extended Data Fig. 4
Pairwise SNP distances for ST405 E. coli.
Source Data Extended Data Fig. 5
Pairwise SNP distances for K. pneumoniae. a, ST11. b, ST15.
Source Data Extended Data Fig. 6
Pairwise SNP distance for ST418 E. cloacae complex.
Source Data Extended Data Fig. 7
(i) Descriptive data for BR, including all samples (A) and samples from the birth cohort (B) and for MR (C) statistical analyses. (ii) Exploratory multivariable analysis performed to understand (A) the association between WASH-related variables and maternal carriage of ARGs using the explanatory variables shown in the table; (B) association between the mother’s handwashing frequency (explanatory variable) and maternal carriage of ARGs (outcome) and controlled for the variables described in the table; (C) association between maternal infection in the 3 months before enrolment in the study (explanatory variable) and maternal carriage of ARGs (outcome) and controlled for the variables shown in the table; (D) association between maternal usage of antibiotics in the 3 months before enrolment in the study (explanatory variable) and maternal carriage of ARGs (outcome) and controlling for the variables depicted in the table. NObs, number of observations. Z-tests were used from multivariable logistic regression models and statistical tests were two sided. For WASH models, P values are adjusted for multiple testing using the Holm–Bonferroni method (FWER = 0.05). (iii) Association between ARG carriage among birth cohort neonates and birth healthcare environment was explored by fitting a logistic regression model to each ARG (outcome) and the explanatory variables shown in the table. The model was re-fitted including PPROM in place of prematurity (these variables were not included in the same model due to their collinear nature). Z-tests were used from multivariable logistic regression models and statistical tests were two sided. P values are adjusted for multiple testing using the Holm–Bonferroni method (FWER = 0.05).
Source Data Table 1
(i) Descriptive data for BRs, including all samples (A) and samples from the birth cohort (B), and for MRs (C) statistical analyses. (ii) Exploratory multivariable analysis performed to understand (A) the association between WASH-related variables and maternal carriage of ARGs using the explanatory variables shown in the table; (B) association between the mother’s handwashing frequency (explanatory variable) and maternal carriage of ARGs (outcome) and controlled for the variables described in the table; (C) association between maternal infection in the 3 months before enrolment in the study (explanatory variable) and maternal carriage of ARGs (outcome) and controlled for the variables shown in the table; (D) association between maternal usage of antibiotics in the 3 months before enrolment in the study (explanatory variable) and maternal carriage of ARGs (outcome) and controlling for the variables depicted in the table. Z-tests were used from multivariable logistic regression models and statistical tests were two sided. For WASH models, P values are adjusted for multiple testing using the Holm–Bonferroni method (FWER = 0.05).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carvalho, M.J., Sands, K., Thomson, K. et al. Antibiotic resistance genes in the gut microbiota of mothers and linked neonates with or without sepsis from low- and middle-income countries. Nat Microbiol 7, 1337–1347 (2022). https://doi.org/10.1038/s41564-022-01184-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-022-01184-y
This article is cited by
-
Duplicated antibiotic resistance genes reveal ongoing selection and horizontal gene transfer in bacteria
Nature Communications (2024)
-
Characterisation of colistin resistance in Gram-negative microbiota of pregnant women and neonates in Nigeria
Nature Communications (2024)
-
Colonisation of hospital surfaces from low- and middle-income countries by extended spectrum β-lactamase- and carbapenemase-producing bacteria
Nature Communications (2024)