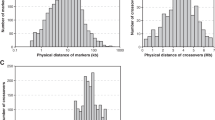
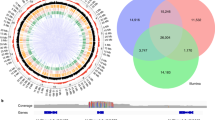
Abstract
GC-favouring gene conversion enables fixation of deleterious alleles, disturbs tests of natural selection and potentially explains both the evolution of recombination as well as the commonly reported intragenomic correlation between G+C content and recombination rate. In addition, gene conversion disturbs linkage disequilibrium, potentially affecting the ability to detect causative variants. However, the importance and generality of these effects is unresolved, not simply because direct analyses are technically challenging but also because previous within- and between-species discrepant results can be hard to appraise owing to methodological differences. Here we report results of methodologically uniform whole-genome sequencing of all tetrad products in Saccharomyces, Neurospora, Chlamydomonas and Arabidopsis. The proportion of polymorphic markers converted varies over three orders of magnitude between species (from 2% of markers converted in yeast to only ~0.005% in the two plants) with at least 87.5% of the variance in per tetrad conversion rates being between species. This is largely due to differences in recombination rate and median tract length. Despite three of the species showing a positive GC-recombination correlation, there is no significant net AT→GC conversion bias in any of the species, despite relatively high resolution in the two taxa (Saccharomyces and Neurospora) with relatively common gene conversion. The absence of a GC bias means that: (1) there should be no presumption that gene conversion is GC biased, or (2) that a GC-recombination correlation necessarily implies biased gene conversion, (3) K a/K s tests should be unaffected in these species and (4) it is unlikely that gene conversion explains the evolution of recombination.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Baudat, F. & de Massy, B. Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosome Res. 15, 565–577 (2007).
Muller, H. J. Some genetic aspects of sex. Am. Nat. 66, 118–138 (1932).
Otto, S. P. & Lenormand, T. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3, 252–261 (2002).
Coop, G. & Przeworski, M. An evolutionary view of human recombination. Nat. Rev. Genet. 8, 23–34 (2007).
Lindegren, C. C. Non-Mendelian segregation in a single tetrad of Saccharomyces ascribed to gene conversion. Science 121, 605–607 (1955).
Mitchell, M. B. Aberrant recombination of pyridoxine mutants of Neurospora. Proc. Natl Acad. Sci. USA 41, 215–220 (1955).
Case, M. E. & Giles, N. H. Evidence from tetrad analysis for both normal and aberrant recombination between allelic mutants in Neurospora crassa. Proc. Natl Acad. Sci. USA 44, 378–390 (1958).
Martinsohn, J. T., Sousa, A. B., Guethlein, L. A. & Howard, J. C. The gene conversion hypothesis of MHC evolution: a review. Immunogenetics 50, 168–200 (1999).
Mondragon-Palomino, M. & Gaut, B. S. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 22, 2444–2456 (2005).
Ardlie, K. et al. Lower-than-expected linkage disequilibrium between tightly linked markers in humans suggests a role for gene conversion. Am. J. Hum. Genet. 69, 582–589 (2001).
Wall, J. D. Close look at gene conversion hot spots. Nat. Genet. 36, 114–115 (2004).
Duret, L. & Galtier, N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10, 285–311 (2009).
Galtier, N., Duret, L., Glémin, S. & Ranwez, V. GC-biased gene conversion promotes the fixation of deleterious amino acid changes in primates. Trends Genet. 25, 1–5 (2009).
Pessia, E. et al. Evidence for widespread GC-biased gene conversion in eukaryotes. Genome Biol. Evol. 4, 675–682 (2012).
Lesecque, Y., Mouchiroud, D. & Duret, L. GC-biased gene conversion in yeast is specifically associated with crossovers: molecular mechanisms and evolutionary significance. Mol. Biol. Evol. 30, 1409–1419 (2013).
Birdsell, J. A. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19, 1181–1197 (2002).
Bengtsson, B. O. Biased conversion as the primary function of recombination. Genet. Res. 47, 77–80 (1986).
Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl Acad. Sci. USA 107, 961–968 (2010).
Hershberg, R. & Petrov, D. A. Evidence that mutation is universally biased towards AT in bacteria. PLOS Genet. 6, e1001115 (2010).
Mancera, E., Bourgon, R., Brozzi, A., Huber, W. & Steinmetz, L. M. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454, 479–485 (2008).
Williams, A. L. et al. Non-crossover gene conversions show strong GC bias and unexpected clustering in humans. eLife 4, e04637 (2015).
Halldorsson, B. V. et al. The rate of meiotic gene conversion varies by sex and age. Nat. Genet. 48, 1377–1384 (2016).
Smeds, L., Mugal, C. F., Qvarnström, A. & Ellegren, H. High-resolution mapping of crossover and non-crossover recombination events by whole-genome re-sequencing of an avian pedigree. PLoS Genet. 12, e1006044 (2016).
Wallberg, A., Glémin, S. & Webster, M. T. Extreme recombination frequencies shape genome variation and evolution in the honeybee, Apis mellifera. PLoS Genet. 11, e1005189 (2015).
Robinson, M. C., Stone, E. A. & Singh, N. D. Population genomic analysis reveals no evidence for GC-biased gene conversion in Drosophila melanogaster. Mol. Biol. Evol. 31, 425–433 (2014).
Odenthal-Hesse, L., Berg, I. L., Veselis, A., Jeffreys, A. J. & May, C. A. Transmission distortion affecting human noncrossover but not crossover recombination: a hidden source of meiotic drive. PLOS Genet. 10, e1004106 (2014).
Berglund, J., Pollard, K. S. & Webster, M. T. Hotspots of biased nucleotide substitutions in human genes. PLOS Biol. 7, e1000026 (2009).
Weber, C. C., Boussau, B., Romiguier, J., Jarvis, E. D. & Ellegren, H. Evidence for GC-biased gene conversion as a driver of between-lineage differences in avian base composition. Genome Biol. 15, 549 (2014).
Eyre-Walker, A. Recombination and mammalian genome evolution. Proc. R. Soc. Lond. B 252, 237–243 (1993).
Marsolier-Kergoat, M.-C. & Yeramian, E. GC content and recombination: reassessing the causal effects for the Saccharomyces cerevisiae genome. Genetics 183, 31–38 (2009).
Jeffreys, A. J. & Neumann, R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum. Mol. Genet. 14, 2277–2287 (2005).
Wijnker, E. et al. The genomic landscape of meiotic crossovers and gene conversions in Arabidopsis thaliana. eLife 2, e01426 (2013).
Cole, F. et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 (2012).
Chicheportiche, A., Bernardino-Sgherri, J., de Massy, B. & Dutrillaux, B. Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J. Cell Sci. 120, 1733–1742 (2007).
Lenzi, M. L. et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am. J. Hum. Genet. 76, 112–127 (2005).
Carofiglio, F. et al. SPO11-independent DNA repair foci and their role in meiotic silencing. PLOS Genet. 9, e1003538 (2013).
Holloway, J. K., Booth, J., Edelmann, W., McGowan, C. H. & Cohen, P. E. MUS81 generates a subset of MLH1–MLH3-independent crossovers in mammalian meiosis. PLOS Genet. 4, e1000186 (2008).
Tease, C., Hartshorne, G. M. & Hultén, M. A. Patterns of meiotic recombination in human fetal oocytes. Am. J. Hum. Genet. 70, 1469–1479 (2002).
Cheng, E. Y. et al. Meiotic recombination in human oocytes. PLoS Genet. 5, e1000661 (2009).
Ottolini, C. S. et al. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47, 727–735 (2015).
Cole, F. et al. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat. Genet. 46, 1072–1080 (2014).
Jeffreys, A. J. & May, C. A. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat. Genet. 36, 151–156 (2004).
Paigen, K. & Petkov, P. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11, 221–233 (2010).
Allers, T. & Lichten, M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57 (2001).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Gerton, J. L. et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 97, 11383–11390 (2000).
Comeron, J. M., Ratnappan, R. & Bailin, S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8, e1002905 (2012).
Liu, H. et al. Causes and consequences of crossing-over evidenced via a high-resolution recombinational landscape of the honey bee. Genome Biol. 16, 15 (2015).
Liu, H. et al. Direct determination of the mutation rate in the bumblebee reveals evidence for weak recombination-associated mutation and an approximate rate constancy in insects. Mol. Biol. Evol. 34, 119–130 (2017).
Yang, S. et al. Parent–progeny sequencing indicates higher mutation rates in heterozygotes. Nature 523, 463–467 (2015).
Charlesworth, B. Genetic recombination: patterns in the genome. Curr. Biol. 4, 182–184 (1994).
Petes, T. D. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2, 360–369 (2001).
Lercher, M. J., Smith, N. G. C., Eyre-Walker, A. & Hurst, L. D. The evolution of isochores: evidence from SNP frequency distributions. Genetics 162, 1805–1810 (2002).
Tsai, I. J., Burt, A. & Koufopanou, V. Conservation of recombination hotspots in yeast. Proc. Natl Acad. Sci. USA 107, 7847–7852 (2010).
Bobay, L.-M. & Ochman, H. Impact of recombination on the base composition of Bacteria and Archaea. Mol. Biol. Evol. 34, 2627–2636 (2017).
Lassalle, F. et al. GC-content evolution in bacterial genomes: the biased gene conversion hypothesis expands. PLOS Genet. 11, e1004941 (2015).
Marais, G., Charlesworth, B. & Wright, S. I. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5, R45 (2004).
Cotton, V. E., Hoffmann, E. R., Abdullah, M. F. F. & Borts, R. H. in Meiosis: Methods in Molecular Biology (Methods and Protocols) Vol. 557 (ed. Keeney, S.) 3–20 (Humana Press, Dordrecht, 2009).
Rockmill, B. et al. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet. 9, e1003932 (2013).
Rockmill, B., Sym, M., Scherthan, H. & Roeder, G. S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 9, 2684–2695 (1995).
Jiang, X. & Stern, D. Mating and tetrad separation of Chlamydomonas reinhardtii for genetic analysis. J. Vis. Exp. 30, e1274 (2009).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Wang, J., Fan, H. C., Behr, B. & Quake, S. R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012).
Oh, S. D. et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 (2007).
Oh, S. D., Lao, J. P., Taylor, A. F., Smith, G. R. & Hunter, N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol. Cell 31, 324–336 (2008).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (91731308, 91631104 and 31671322 to S.Y. or D.T.) and the European Research Council (Advanced grant ERC-2014-ADG 669207 to L.D.H.). We are indebted to D. Liu for advice on Chlamydomonas experiments. We thank S. Li, C. Tian and W. Sun for providing the Neurospora strains that were used in this study.
Author information
Authors and Affiliations
Contributions
S.Y., L.D.H., D.T. and H.L. designed the experiments. H.L., J.H., X.S., J.L., Y.H., L.Y., S.Y, L.D.H. and G.L. performed the experiments and analysed the data. L.D.H., H.L., S.Y. and D.T. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary discussion; supplementary figures 1–11; supplementary tables 1–7.
Rights and permissions
About this article
Cite this article
Liu, H., Huang, J., Sun, X. et al. Tetrad analysis in plants and fungi finds large differences in gene conversion rates but no GC bias. Nat Ecol Evol 2, 164–173 (2018). https://doi.org/10.1038/s41559-017-0372-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0372-7
This article is cited by
-
Selection on synonymous sites: the unwanted transcript hypothesis
Nature Reviews Genetics (2024)
-
Loss of Heterozygosity and Its Importance in Evolution
Journal of Molecular Evolution (2023)