Abstract
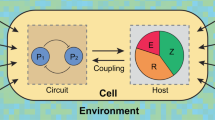
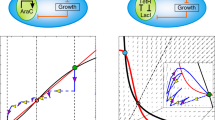
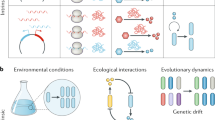
One fundamental challenge in synthetic biology is the lack of quantitative tools that accurately describe and predict the behaviours of engineered gene circuits. This challenge arises from multiple factors, among which the complex interdependence of circuits and their host is a leading cause. Here we present a gene circuit modelling framework that explicitly integrates circuit behaviours with host physiology through bidirectional circuit–host coupling. The framework consists of a coarse-grained but mechanistic description of host physiology that involves dynamic resource partitioning, multilayered circuit–host coupling including both generic and system-specific interactions, and a detailed kinetic module of exogenous circuits. We showed that, following training, the framework was able to capture and predict a large set of experimental data concerning the host and its foreign gene overexpression. To demonstrate its utility, we applied the framework to examine a growth-modulating feedback circuit whose dynamics is qualitatively altered by circuit–host interactions. Using an extended version of the framework, we further systematically revealed the behaviours of a toggle switch across scales from single-cell dynamics to population structure and to spatial ecology. This work advances our quantitative understanding of gene circuit behaviours and also benefits the rational design of synthetic gene networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000).
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000).
Basu, S., Gerchman, Y., Collins, C. H., Arnold, F. H. & Weiss, R. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130–1134 (2005).
Elowitz, M. & Lim, W. A. Build life to understand it. Nature 468, 889–890 (2010).
Tabor, J. J. et al. A synthetic genetic edge detection program. Cell 137, 1272–1281 (2009).
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B. & Keasling, J. D. Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012).
Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nat. Rev. Genet. 11, 367–379 (2010).
Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science 333, 1248–1252 (2011).
Lu, T. K., Khalil, A. S. & Collins, J. J. Next-generation synthetic gene networks. Nat. Biotechnol. 27, 1139–1150 (2009).
Kwok, R. Five hard truths for synthetic biology. Nature 463, 288–290 (2010).
Brophy, J. A. & Voigt, C. A. Principles of genetic circuit design. Nat. Methods 11, 508–520 (2014).
Cardinale, S. & Arkin, A. P. Contextualizing context for synthetic biology–identifying causes of failure of synthetic biological systems. Biotechnol. J. 7, 856–866 (2012).
Arkin, A. P. A wise consistency: engineering biology for conformity, reliability, predictability. Curr. Opin. Chem. Biol. 17, 893–901 (2013).
Zhang, C., Tsoi, R. & You, L. Addressing biological uncertainties in engineering gene circuits. Integr. Biol. 8, 456–464 (2016).
Venturelli, O. S. et al. Programming mRNA decay to modulate synthetic circuit resource allocation. Nat. Commun. 8, 15128 (2017).
Potrykus, K., Murphy, H., Philippe, N. & Cashel, M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol 13, 563–575 (2011).
Traxler, M. F. et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148 (2008).
Potrykus, K. & Cashel, M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51 (2008).
Anderson, J. C., Voigt, C. A. & Arkin, A. P. Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 3, 133 (2007).
Deris, J. B. et al. The innate growth bistability and fitness landscapes of antibiotic-resistant bacteria. Science 342, 1237435 (2013).
Glick, B. R. Metabolic load and heterologous gene expression. Biotechnol. Adv. 13, 247–261 (1995).
Tan, C., Marguet, P. & You, L. Emergent bistability by a growth-modulating positive feedback circuit. Nat. Chem. Biol. 5, 842–848 (2009).
Klumpp, S., Zhang, Z. & Hwa, T. Growth rate-dependent global effects on gene expression in bacteria. Cell 139, 1366–1375 (2009).
Karr, J. R. et al. A whole-cell computational model predicts phenotype from genotype. Cell 150, 389–401 (2012).
Scott, M., Gunderson, C. W., Mateescu, E. M., Zhang, Z. & Hwa, T. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010).
Weiße, A. Y., Oyarzún, D. A., Danos, V. & Swain, P. S. Mechanistic links between cellular trade-offs, gene expression, and growth. Proc. Natl Acad. Sci. USA 112, E1038–E1047 (2015).
Maitra, A. & Dill, K. A. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc. Natl Acad. Sci. USA 112, 406–411 (2015).
Marr, A. G. Growth rate of Escherichia coli. Microbiol. Rev. 55, 316–333 (1991).
Gerosa, L., Kochanowski, K., Heinemann, M. & Sauer, U. Dissecting specific and global transcriptional regulation of bacterial gene expression. Mol. Syst. Biol. 9, 658 (2013).
Paul, B. J. et al. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118, 311–322 (2004).
Hartl, F. U., Bracher, A. & Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011).
Li, G.-W., Burkhardt, D., Gross, C. & Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014).
Lu, T., Shen, T., Bennett, M. R., Wolynes, P. G. & Hasty, J. Phenotypic variability of growing cellular populations. Proc. Natl Acad. Sci. USA 104, 18982–18987 (2007).
Blanchard, A. E. & Lu, T. Bacterial social interactions drive the emergence of differential spatial colony structures. BMC Syst. Biol. 9, 59 (2015).
Gillespie, D. T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361 (1977).
Bremer, H. & Dennis, P. P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus https://doi.org/10.1128/ecosal.5.2.3 (2008).
Klumpp, S. & Hwa, T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl Acad. Sci. USA 105, 20245–20250 (2008).
Bollenbach, T., Quan, S., Chait, R. & Kishony, R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139, 707–718 (2009).
Friesen, J. D., Fiil, N. & Von Meyenburg, K. Synthesis and turnover of basal level guanosine tetraphosphate in Escherichia coli. J. Biol. Chem. 250, 304–309 (1975).
Koch, A. L. & Deppe, C. S. In vivo assay of protein synthesizing capacity of Escherichia coli from slowly growing chemostat cultures. J. Mol. Biol. 55, 549–562 (1971).
Molin, S., Von Meyenburg, K., Maaløe, O., Hansen, M. T. & Pato, M. L. Control of ribosome synthesis in Escherichia coli: analysis of an energy source shift-down. J. Bacteriol. 131, 7–17 (1977).
Johnsen, K., Molin, S., Karlström, O. & Maaløe, O. Control of protein synthesis in Escherichia coli: analysis of an energy source shift-down. J. Bacteriol. 131, 18–29 (1977).
Barker, M. M., Gaal, T., Josaitis, C. A. & Gourse, R. L. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J. Mol. Biol. 305, 673–688 (2001).
Dong, H., Nilsson, L. & Kurland, C. G. Gratuitous overexpression of genes in Escherichia coli leads to growth inhibition and ribosome destruction. J. Bacteriol. 177, 1497–1504 (1995).
Murray, H. D., Schneider, D. A. & Gourse, R. L. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12, 125–134 (2003).
Chopra, I., Hacker, K., Misulovin, Z. & Rothstein, D. Sensitive biological detection method for tetracyclines using a tetA-lacZ fusion system. Antimicrob. Agents Chemother. 34, 111–116 (1990).
Schaechter, M., Maaløe, O. & Kjeldgaard, N. O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology 19, 592–606 (1958).
Karpinets, T. V., Greenwood, D. J., Sams, C. E. & Ammons, J. T. RNA:protein ratio of the unicellular organism as a characteristic of phosphorous and nitrogen stoichiometry and of the cellular requirement of ribosomes for protein synthesis. BMC Biol. 4, 30 (2006).
Cox, R. A. Quantitative relationships for specific growth rates and macromolecular compositions of Mycobacterium tuberculosis, Streptomyces coelicolor A3 (2) and Escherichia coli B/r: an integrative theoretical approach. Microbiology 150, 1413–1426 (2004).
Tozaki, H. et al. Reconstructing the single-cell-level behavior of a toggle switch from population-level measurements. FEBS Lett. 582, 1067–1072 (2008).
Acknowledgements
This work was supported by the National Science Foundation (No. 1553649 and 1227034), the Office of Naval Research (No. N000141612525), the American Heart Association (No. 12SDG12090025), the Brain and Behavior Research Foundation (NARSAD Young Investigator Award), and the National Center for Supercomputing Applications (Faculty Fellowship).
Author information
Authors and Affiliations
Contributions
T. L. conceived and directed the study and designed the research. C.L. and A.E.B. contributed to model development. C.L., A.E.B. and T.L. analysed data. T.L. and C.L. wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Methods, Supplementary Tables 1–4, Supplementary Figures 1–32, Supplementary References.
Supplementary Information
Life Sciences Reporting Summary
Rights and permissions
About this article
Cite this article
Liao, C., Blanchard, A.E. & Lu, T. An integrative circuit–host modelling framework for predicting synthetic gene network behaviours. Nat Microbiol 2, 1658–1666 (2017). https://doi.org/10.1038/s41564-017-0022-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-017-0022-5
This article is cited by
-
A coarse-grained bacterial cell model for resource-aware analysis and design of synthetic gene circuits
Nature Communications (2024)
-
Modelling genetic stability in engineered cell populations
Nature Communications (2023)
-
Resource-aware whole-cell model of division of labour in a microbial consortium for complex-substrate degradation
Microbial Cell Factories (2022)
-
Dynamic cybergenetic control of bacterial co-culture composition via optogenetic feedback
Nature Communications (2022)
-
Understanding and mathematical modelling of cellular resource allocation in microorganisms: a comparative synthesis
BMC Bioinformatics (2021)