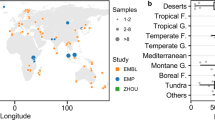
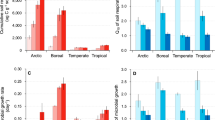
The legacy impacts of past climates on the current distribution of soil microbial communities are largely unknown. Here, we use data from more than 1,000 sites from five separate global and regional datasets to identify the importance of palaeoclimatic conditions (Last Glacial Maximum and mid-Holocene) in shaping the current structure of soil bacterial communities in natural and agricultural soils. We show that palaeoclimate explains more of the variation in the richness and composition of bacterial communities than current climate. Moreover, palaeoclimate accounts for a unique fraction of this variation that cannot be predicted from geographical location, current climate, soil properties or plant diversity. Climatic legacies (temperature and precipitation anomalies from the present to ~20 kyr ago) probably shape soil bacterial communities both directly and indirectly through shifts in soil properties and plant communities. The ability to predict the distribution of soil bacteria from either palaeoclimate or current climate declines greatly in agricultural soils, highlighting the fact that anthropogenic activities have a strong influence on soil bacterial diversity. We illustrate how climatic legacies can help to explain the current distribution of soil bacteria in natural ecosystems and advocate that climatic legacies should be considered when predicting microbial responses to climate change.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Monger, C. et al. Legacy effects in linked ecological–soil–geomorphic systems of drylands. Front. Ecol. Environ. 13, 13–19 (2015).
Gajewski, K. Impact of Holocene climate variability on Arctic vegetation. Glob. Planet. Chang. 133, 272–287 (2015).
Svenning, J.-C. et al. The influence of paleoclimate on present-day patterns in biodiversity and ecosystems. Annu. Rev. Ecol. Evol. 46, 551–572 (2015).
Lyons, S. K. et al. Holocene shifts in the assembly of plant and animal communities implicate human impacts. Nature 529, 80–83 (2016).
Martiny, J. B. H. History leaves its mark on soil bacterial diversity. mBio 7, e00784-16 (2016).
Andam, C. P. et al. A latitudinal diversity gradient in terrestrial bacteria of the genus Streptomyces. mBio 7, e02200-15 (2016).
Wagg, C. et al. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl Acad. Sci. USA 111, 5266–5270 (2014).
Delgado-Baquerizo, M. et al. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 28, 10541 (2016).
van der Heijden, M. G. et al. The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008).
Oliverio, A. et al. Identifying the microbial taxa that consistently respond to soil warming across time and space. Glob. Chang. Biol. 23, 2117–2129 (2017).
Atkinson, T. C. et al. Seasonal temperatures in Britain during the past 22,000 years, reconstructed using beetle remains. Nature 325, 587–592 (1987).
Fordham, D. A. et al. PaleoView: a tool for generating continuous climate projections spanning the last 21000 years at regional and global scales. Ecography http://dx.doi.org/10.1111/ecog.03031 (2017).
Lauber, C. L. et al. Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment. Appl. Environ. Microbiol. 75, 5111–5120 (2009).
Delgado-Baquerizo, M. et al. Carbon content and climate variability drive global soil bacterial diversity patterns. Ecol. Monogr. 86, 373–390 (2016).
Maestre, F. T. et al. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl Acad. Sci. USA 112, 15684–15689 (2015).
Vitousek, P. M. Nutrient Cycling and Limitation: Hawai’i as a Model System (Princeton University, New Jersey, 2004).
Wardle, D. A. et al. Ecosystem properties and forest decline in contrasting long-term chronosequences. Science 305, 509–513 (2004).
Prober, S. M. et al. Plant diversity predicts beta but not alpha diversity of soil microbes across grasslands worldwide. Ecol. Lett. 18, 85–95 (2015).
Leff, J. W. et al. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 214, 412–423 (2017).
Leff, J. W. et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl Acad. Sci. USA 112, 10967–10972 (2015).
Trivedi, P. et al. Response of soil properties and microbial communities to agriculture: implications for primary productivity and soil health indicators. Front. Plant Sci. 7, 990 (2016).
World Development Report: Agriculture for Development (World Bank, Washington DC, 2008).
Legendre, P. et al. Studying beta diversity: ecological variation partitioning by multiple regression and canonical analysis. J. Plant Ecol. 1, 3–8 (2008).
Nogués-Bravo, D. et al. Amplified plant turnover in response to climate change forecast by Late Quaternary records. Nat. Clim. Chang. 6, 1115–1119 (2016).
Fieseler, L. et al. Discovery of the novel candidate phylum ‘Poribacteria’ in marine sponges. Appl. Environ. Microbiol. 70, 3724–3732 (2004).
Youssef, N. H. et al. In silico analysis of the metabolic potential and niche specialization of candidate phylum ‘Latescibacteria’ (WS3). PLoS ONE 10, e0127499 (2015).
Schlesinger, W. H. & Bernhardt, E. S. Biogeochemistry: an Analysis of Global Change (Academic Press, San Diego, 1996).
Ramirez, K. S. et al. Biogeographic patterns in belowground diversity in New York City’s Central Park are similar to those observed globally. Proc. R. Soc. Lond. B Biol. Sci. 281, 20141988 (2014).
Wang, J.-T. et al. Coupling of soil prokaryotic diversity and plant diversity across latitudinal forest ecosystems. Sci. Rep. 5, 19561 (2015).
United Nations Environment Programme World Atlas of Desertification (Edward Arnold, London, 2002).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J. 5, 1571–1579 (2011).
Maestre, F. T. et al. Plant species richness and ecosystem multifunctionality in global drylands. Science 335, 214–218 (2012).
Bates, S. et al. Bacterial communities associated with the lichen symbiosis. Appl. Environ. Microbiol. 77, 1309–1314 (2011).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Bissett, A. et al. Introducing BASE: the Biomes of Australian Soil Environments soil microbial diversity database. GigaScience 20165, 21 (2016).
Lane, D. J. Nucleic Acid Techniques in Bacterial Systematics (Wiley, New York, 1991).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol 75, 7537–7541 (2009).
Eldridge, D. J. et al. Competition drives the response of soil microbial diversity to increased grazing by vertebrate herbivores. Ecology 98, 1922–1931 (2017).
Edgar, R. G. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Gent, P. R. et al. The Community Climate System Model version 4. J. Clim. 24, 4973–4991 (2011).
Bystriakova, N. et al. Present, past and future of the European rock fern Asplenium fontanum: combining distribution modelling and population genetics to study the effect of climate change on geographic range and genetic diversity. Ann. Bot. 13, 453–465 (2013).
Tallavaaraa, M. et al. Human population dynamics in Europe over the Last Glacial Maximum. Proc. Natl Acad. Sci. USA 112, 8232–8237 (2015).
Delgado-Baquerizo, M. et al. Biogeographic bases for a shift in crop C:N:P stoichiometries during domestication. Ecol. Lett. 19, 564–575 (2016).
Katz, M. H. Multivariable Analysis: A Practical Guide for Clinicians and Public Health Researchers (Cambridge Univ. Press, Cambridge, 2006).
Oksanen, J. et al. vegan: Community Ecology Package. R package v. 2.3-0 (2015).
Breiman, L. Random forests. Mach. Learn. 45, 5 (2001).
Archer, E. rfPermute: Estimate Permutation p-Values for Random Forest Importance Metrics. R package v. 1.5.2 (2016).
Grace, J. B. Structural Equation Modeling Natural Systems (Cambridge Univ. Press, Cambridge, 2006).
Schermelleh-Engel, K. et al. Evaluating the fit of structural equation models: tests of significance and descriptive goodness-of-fit measures. Methods Psychol. Res. 8, 23–74 (2003).
Delgado-Baquerizo, M. et al. Lack of functional redundancy in the relationship between microbial diversity and ecosystem functioning. J. Ecol. 104, 936–946 (2016).
Acknowledgements
M.D.-B. acknowledges support from the Marie Sklodowska-Curie Actions of the Horizon 2020 Framework Programme H2020-MSCA-IF-2016 under REA grant agreement no. 702057. We acknowledge the contribution of the BASE project partners (DOI: 10.4227/71/561c9bc670099), an initiative supported by Bioplatforms Australia with funds provided by the Australian Commonwealth Government through the National Collaborative Research Infrastructure Strategy. B.K.S. and M.D-B are supported by the Australian Research Council projects (DP13010484 and DP170104634). D.J.E. was supported by the Hermon Slade Foundation. N.F was supported by grants from the US National Science Foundation (PLR 1241629 and DEB 1542653). The work from J.-Z.H. and the China dataset were supported by the Natural Science Foundation of China (grant no. 41230857) and the Chinese Academy of Sciences (grant no. XDB15020200). The work of F.T.M. and the Global Drylands database were supported by the European Research Council (ERC grant agreements 242658 [BIOCOM] and 647038 [BIODESERT]).
Author information
Authors and Affiliations
Contributions
M.D.-B. conceived the idea of this study in consultation with N.F. The microbial datasets of the Global Drylands were originally compiled by F.T.M, B.K.S. and M.D.-B.; those of the Americas by N.F.; Australia by A.B.; China by J.-Z.H., Y.-R.L. and J.-T.W.; and New South Wales by D.J.E., B.K.S. and K.H. Statistical modelling was conducted by M.D.-B. The manuscript was written by M.D.-B. with contributions from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Correspondence and requests for materials should be addressed to M.D.-B.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Appendices 1–3, Supplementary Tables 1, 3, 4, 8–12, Supplementary Figures 1–15
Supplementary_Table_2
Correlation (Pearson) among bioclimatic variables across different time periods. Worldclim number of climatic variables are shown in Supplementary Table 1
Supplementary_Table_5
Standardized direct effects from s.e.m. in Fig. 2 and Supplementary Figs. 6 and 7
Supplementary_Table_6
Correlations (standardized effects) from s.e.m. in Fig. 2 and Supplementary Figs. 6 and 7
Supplementary_Table_7
Results from random forest analyses aiming to identify the most important bacterial composition predictors of selected palaeoclimatic legacies (AMT or PDM). Acronyms of climatic variables are shown in Supplementary Table 1. Importance is calculated as the per cent increase in the mean square error in our models
Rights and permissions
About this article
Cite this article
Delgado-Baquerizo, M., Bissett, A., Eldridge, D.J. et al. Palaeoclimate explains a unique proportion of the global variation in soil bacterial communities. Nat Ecol Evol 1, 1339–1347 (2017). https://doi.org/10.1038/s41559-017-0259-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0259-7
This article is cited by
-
Enzyme adaptation to habitat thermal legacy shapes the thermal plasticity of marine microbiomes
Nature Communications (2023)
-
Climate legacies drive the distribution and future restoration potential of dryland forests
Nature Plants (2022)
-
The neglected role of micronutrients in predicting soil microbial structure
npj Biofilms and Microbiomes (2022)
-
Past climate conditions predict the influence of nitrogen enrichment on the temperature sensitivity of soil respiration
Communications Earth & Environment (2021)
-
Ecosystem properties in urban areas vary with habitat type and settlement age
Plant and Soil (2021)