Abstract
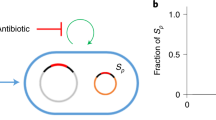
Horizontal gene transfer (HGT) between bacterial lineages is a fundamental evolutionary process that accelerates adaptation. Sequence analyses show that conjugative plasmids are principal agents of HGT in natural communities. However, we lack understanding of how the ecology of bacterial communities and their environments affect the dynamics of plasmid-mediated gene mobilization and transfer. Here we show, in simple experimental soil bacterial communities containing a conjugative mercury resistance plasmid, the repeated, independent mobilization of transposon-borne genes from chromosome to plasmid, plasmid to chromosome and, in the absence of mercury selection, interspecific gene transfers from the chromosome of one species to the other via the plasmid. By reducing conjugation, positive selection for plasmid-encoded traits, like mercury resistance, can consequently inhibit HGT. Our results suggest that interspecific plasmid-mediated gene mobilization is most likely to occur in environments where plasmids are infectious, parasitic elements rather than those where plasmids are positively selected, beneficial elements.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Halary, S., Leigh, J. W., Cheaib, B., Lopez, P. & Bapteste, E. Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl Acad. Sci. USA 107, 127–132 (2010).
Norman, A., Hansen, L. H. & Sorensen, S. J. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275–2289 (2009).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (2005).
Conlan, S. et al. Plasmid dynamics in KPC-positive Klebsiella pneumoniae during long-term patient colonization. mBio 7, e00742-16 (2016).
Sheppard, A. E. et al. Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene bla KPC. Antimicrob. Agents Chemother. 60, 3767–3778 (2016).
Rasmussen, S. et al. Early divergent strains of Yersinia pestis in Eurasia 5,000 years ago. Cell 163, 571–582 (2015).
Johnson, T. J. & Nolan, L. K. Pathogenomics of the virulence plasmids of Escherichia coli. Microbiol. Mol. Biol. Rev. 73, 750–774 (2009).
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2002).
Lopatkin, A. J. et al. Antibiotics as a selective driver for conjugation dynamics. Nat. Microbiol. 1, 16044 (2016).
Stevenson, C., Hall, J. P., Harrison, E., Wood, A. J. & Brockhurst, M. A. Gene mobility promotes the spread of resistance in bacterial populations. ISME J. 63, 1577 (2017).
Jacoby, G. A., Rogers, J. E., Jacob, A. E. & Hedges, R. W. Transposition of Pseudomonas toluene-degrading genes and expression in Escherichia coli. Nature 274, 179–180 (1978).
Hedges, R. W. & Jacob, A. E. In vivo translocation of genes of Pseudomonas aeruginosa onto a promiscuously transmissible plasmid. FEMS Microbiol. Lett. 2, 15–19 (1977).
Hemme, C. L. et al. Lateral gene transfer in a heavy metal-contaminated-groundwater microbial community. mBio 7, e02234-15 (2016).
Xue, H. et al. Eco-evolutionary dynamics of episomes among ecologically cohesive bacterial populations. mBio 6, e00552-15 (2015).
Gomez, P. & Buckling, A. Bacteria-phage antagonistic coevolution in soil. Science 332, 106–109 (2011).
Hall, J. P. J. et al. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 17, 5008–5022 (2015).
Hall, J. P. J., Wood, A. J., Harrison, E. & Brockhurst, M. A. Source–sink plasmid transfer dynamics maintain gene mobility in soil bacterial communities. Proc. Natl Acad. Sci. USA 113, 8260–8265 (2016).
Li, P., Feng, X. B., Qiu, G. L., Shang, L. H. & Li, Z. G. Mercury pollution in Asia: a review of the contaminated sites. J. Hazard. Mater. 168, 591–601 (2009).
Williams, D., Paterson, S., Brockhurst, M. A. & Winstanley, C. Refined analyses suggest that recombination is a minor source of genomic diversity in Pseudomonas aeruginosa chronic cystic fibrosis infections. Microb. Genom. 2, e000051 (2016).
Silby, M. W. et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 10, R51 (2009).
Ravindran, A., Jalan, N., Yuan, J. S., Wang, N. & Gross, D. C. Comparative genomics of Pseudomonas syringae pv. syringae strains B301D and HS191 and insights into intrapathovar traits associated with plant pathogenesis. MicrobiologyOpen 4, 553–573 (2015).
Kivistik, P. A., Kivisaar, M. & Horak, R. Target site selection of Pseudomonas putida transposon Tn4652. J. Bacteriol. 189, 3918–3921 (2007).
Levin, B. R., Stewart, F. M. & Rice, V. A. The kinetics of conjugative plasmid transmission: fit of a simple mass action model. Plasmid 2, 247–260 (1979).
Hausner, M. & Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65, 3710–3713 (1999).
Bergstrom, C. T., Lipsitch, M. & Levin, B. R. Natural selection, infectious transfer and the existence conditions for bacterial plasmids. Genetics 155, 1505–1519 (2000).
Harrison, E. et al. Rapid compensatory evolution promotes the survival of conjugative plasmids. Mob. Genet. Elements 6, e1179074 (2016).
Baltrus, D. A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013).
McCarthy, A. J. et al. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol. Evol. 6, 2697–2708 (2014).
Shapiro, B. J. How clonal are bacteria over time? Curr. Opin. Microbiol. 31, 116–123 (2016).
Polz, M. F., Alm, E. J. & Hanage, W. P. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet. 29, 170–175 (2013).
Sousa, A., Bourgard, C., Wahl, L. M. & Gordo, I. Rates of transposition in Escherichia coli. Biol. Lett. 9, 20130838 (2013).
Dennis, J. J. The evolution of IncP catabolic plasmids. Curr. Opin. Biotechnol. 16, 291–298 (2005).
Revilla, C. et al. Different pathways to acquiring resistance genes illustrated by the recent evolution of IncW plasmids. Antimicrob. Agents Chemother. 52, 1472–1480 (2008).
Carraro, N., Rivard, N., Burrus, V. & Ceccarelli, D. Mobilizable genomic islands, different strategies for the dissemination of multidrug resistance and other adaptive traits. Mob. Genet. Elements 7, 1–6 (2017).
Condit, R., Stewart, F. M. & Levin, B. R. The population biology of bacterial transposons: a priori conditions for maintenance as parasitic DNA. Am. Nat. 132, 129–147 (1988).
Twiss, E., Coros, A. M., Tavakoli, N. P. & Derbyshire, K. M. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol. Microbiol. 57, 1593–1607 (2005).
Baharoglu, Z., Bikard, D. & Mazel, D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 6, e1001165 (2010).
Christie-Oleza, J. A., Lanfranconi, M. P., Nogales, B., Lalucat, J. & Bosch, R. Conjugative interaction induces transposition of ISPst9 in Pseudomonas stutzeri AN10. J. Bacteriol. 191, 1239–1247 (2009).
He, S. et al. Mechanisms of evolution in high-consequence drug resistance plasmids. mBio 7, e01987-16 (2016).
O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations (Wellcome Trust & HM Government, London, 2016).
Cho, J. C. & Tiedje, J. M. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66, 5448–5456 (2000).
De Gelder, L., Williams, J. J., Ponciano, J. M., Sota, M. & Top, E. M. Adaptive plasmid evolution results in host-range expansion of a broad-host-range plasmid. Genetics 178, 2179–2190 (2008).
Harrison, E., Guymer, D., Spiers, A. J., Paterson, S. & Brockhurst, M. A. Parallel compensatory evolution stabilizes plasmids across the parasitism–mutualism continuum. Curr. Biol. 25, 2034–2039 (2015).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681 (2009).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Lilley, A. K., Bailey, M. J., Day, M. J. & Fry, J. C. Diversity of mercury resistance plasmids obtained by exogenous isolation from the bacteria of sugar beet in three successive years. FEMS Microbiol. Ecol. 20, 211–227 (1996).
Acknowledgements
We thank P. Koldkjaer and others at the Liverpool Centre for Genomic Research for assistance with sample preparation and sequencing. This work was supported by ERC Grant Agreement no. 311490-COEVOCON to M.A.B. and a Philip Leverhulme Prize from Leverhulme Trust to M.A.B.
Author information
Authors and Affiliations
Contributions
J.P.J.H., E.H. and M.A.B. designed the study; J.P.J.H. collected data; J.P.J.H., D.W. and S.P. analysed the data. J.P.J.H. and M.A.B. drafted the manuscript. All authors discussed results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Tables 1–4
Rights and permissions
About this article
Cite this article
Hall, J.P.J., Williams, D., Paterson, S. et al. Positive selection inhibits gene mobilization and transfer in soil bacterial communities. Nat Ecol Evol 1, 1348–1353 (2017). https://doi.org/10.1038/s41559-017-0250-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0250-3
This article is cited by
-
Intra- and interpopulation transposition of mobile genetic elements driven by antibiotic selection
Nature Ecology & Evolution (2022)
-
Plasmid- and strain-specific factors drive variation in ESBL-plasmid spread in vitro and in vivo
The ISME Journal (2021)
-
Beyond horizontal gene transfer: the role of plasmids in bacterial evolution
Nature Reviews Microbiology (2021)
-
The persistence potential of transferable plasmids
Nature Communications (2020)
-
Can mobile genetic elements rescue genes from extinction?
Current Genetics (2020)