Key Points
-
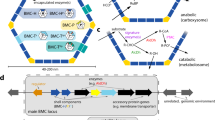
Bacterial microcompartments are functional analogues of the lipid-bound organelles of eukaryotes. They enclose chemical reactions that benefit from being separated from the cytosol.
-
The delimiting membrane of bacterial microcompartments consists entirely of protein, and its components are highly conserved in sequence and structure.
-
Bacterial microcompartments are found in a wide variety of bacterial species (at least 19 established phyla). They are easily identified in genomes by their tendency to colocalize the associated genes into a large gene cluster called a superlocus.
-
Carboxysomes (CO2-fixing organelles) were the first type of bacterial microcompartment to be identified, but recently, many more have been discovered and characterized; they are involved in catabolizing a variety of nutrients and enable cells to grow in otherwise unavailable niches.
-
The shell and cargo of bacterial microcompartments self-assemble using different pathways; some build the shell around a cargo aggregate, whereas others assemble the shell and cargo concomitantly. There are proteins that facilitate cargo aggregation and small encapsulation peptides that specifically associate proteins to the lumen of the shell.
-
Bacterial microcompartments are linked to the pathogenesis of certain bacteria because they confer a growth advantage. For example, the human gut is enriched in propanediol and ethanolamine, initial substrates of specific bacterial microcompartments.
-
The knowledge gained from understanding the native functions has led to substantial progress in modifying the shell for bioengineering purposes. Bacterial microcompartment shells can be produced recombinantly, and shell proteins and cores have been engineered to adopt new functions.
Abstract
Bacterial microcompartments (BMCs) are self-assembling organelles that consist of an enzymatic core that is encapsulated by a selectively permeable protein shell. The potential to form BMCs is widespread and found across the kingdom Bacteria. BMCs have crucial roles in carbon dioxide fixation in autotrophs and the catabolism of organic substrates in heterotrophs. They contribute to the metabolic versatility of bacteria, providing a competitive advantage in specific environmental niches. Although BMCs were first visualized more than 60 years ago, it is mainly in the past decade that progress has been made in understanding their metabolic diversity and the structural basis of their assembly and function. This progress has not only heightened our understanding of their role in microbial metabolism but is also beginning to enable their use in a variety of applications in synthetic biology. In this Review, we focus on recent insights into the structure, assembly, diversity and function of BMCs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kerfeld, C. A., Heinhorst, S. & Cannon, G. C. Bacterial microcompartments. Annu. Rev. Microbiol. 64, 391–408 (2010).
Kerfeld, C. A. & Melnicki, M. R. Assembly, function and evolution of cyanobacterial carboxysomes. Curr. Opin. Plant Biol. 31, 66–75 (2016).
Rae, B. D., Long, B. M., Badger, M. R. & Price, G. D. Functions, compositions, and evolution of the two types of carboxysomes: polyhedral microcompartments that facilitate CO2 fixation in cyanobacteria and some proteobacteria. Microbiol. Mol. Biol. Rev. 77, 357–379 (2013).
Turmo, A., Gonzalez-Esquer, C. R. & Kerfeld, C. A. Carboxysomes: metabolic modules for CO2 fixation. FEMS Microbiol. Lett. 364, fnx176 (2017).
Chowdhury, C., Sinha, S., Chun, S., Yeates, T. O. & Bobik, T. A. Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 78, 438–468 (2014).
Axen, S. D., Erbilgin, O. & Kerfeld, C. A. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLOS Comput. Biol. 10, e1003898 (2014). This is a comprehensive survey of BMC types and subtypes by bioinformatic analysis of superloci.
Kerfeld, C. A. & Erbilgin, O. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol. 23, 22–34 (2015).
Zarzycki, J., Erbilgin, O. & Kerfeld, C. A. Bioinformatic characterization of glycyl radical enzyme-associated bacterial microcompartments. Appl. Environ. Microbiol. 81, 8315–8329 (2015).
Jakobson, C. M., Tullman-Ercek, D., Slininger, M. F. & Mangan, N. M. A systems-level model reveals that 1,2-Propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLOS Comput. Biol. 13, e1005525 (2017).
Cannon, G. C., Heinhorst, S. & Kerfeld, C. A. Carboxysomal carbonic anhydrases: structure and role in microbial CO2 fixation. Biochim. Biophys. Acta 1804, 382–392 (2010).
Bobik, T. A., Havemann, G. D., Busch, R. J., Williams, D. S. & Aldrich, H. C. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181, 5967–5975 (1999).
Kofoid, E., Rappleye, C., Stojiljkovic, I. & Roth, J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181, 5317–5329 (1999).
Petit, E. et al. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLOS ONE 8, e54337 (2013). This study presents the first experimental characterization of a GRM organelle.
Erbilgin, O., McDonald, K. L. & Kerfeld, C. A. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl. Environ. Microbiol. 80, 2193–2205 (2014). This study reports the functional characterization of a PVM BMC involved in the degradation of fucose and rhamnose.
Roof, D. M. & Roth, J. R. Autogenous regulation of ethanolamine utilization by a transcriptional activator of the eut operon in Salmonella typhimurium. J. Bacteriol. 174, 6634–6643 (1992).
Bobik, T. A., Ailion, M. & Roth, J. R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174, 2253–2266 (1992).
Jakobson, C. M. & Tullman-Ercek, D. Dumpster diving in the gut: bacterial microcompartments as part of a host-associated lifestyle. PLOS Pathog. 12, e1005558 (2016).
Yeates, T. O., Kerfeld, C. A., Heinhorst, S., Cannon, G. C. & Shively, J. M. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6, 681–691 (2008).
Sutter, M. et al. Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy. Nano Lett. 16, 1590–1595 (2016).
Sutter, M., Greber, B., Aussignargues, C. & Kerfeld, C. A. Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297 (2017). This study shows that the structure of a complete BMC shell reveals specific interactions among shell proteins.
Savage, D. F., Afonso, B., Chen, A. H. & Silver, P. A. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 327, 1258–1261 (2010). This study shows alignment of β-carboxysomes along the long axis of S. elongatus by fluorescent labelling and correlates this to interaction with the cytoskeleton.
Menon, B. B., Heinhorst, S., Shively, J. M. & Cannon, G. C. The carboxysome shell is permeable to protons. J. Bacteriol. 192, 5881–5886 (2010).
Parsons, J. B. et al. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol. Cell 38, 305–315 (2010).
Frank, S., Lawrence, A. D., Prentice, M. B. & Warren, M. J. Bacterial microcompartments moving into a synthetic biological world. J. Biotechnol. 163, 273–279 (2013).
Gonzalez-Esquer, C. R., Newnham, S. E. & Kerfeld, C. A. Bacterial microcompartments as metabolic modules for plant synthetic biology. Plant J. 87, 66–75 (2016).
Plegaria, J. S. & Kerfeld, C. A. Engineering nanoreactors using bacterial microcompartment architectures. Curr. Opin. Biotechnol. 51, 1–7 (2017).
Kerfeld, C. A. et al. Protein structures forming the shell of primitive bacterial organelles. Science 309, 936–938 (2005).
Cai, F. et al. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J. Biol. Chem. 288, 16055–16063 (2013).
Klein, M. G. et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392, 319–333 (2009). This study reports the first structure of a double stacking BMC-T protein and a possible pore gating mechanism.
Mallette, E. & Kimber, M. S. A. Complete structural inventory of the mycobacterial microcompartment shell proteins constrains models of global architecture and transport. J. Biol. Chem. 292, 1197–1210 (2017).
Larsson, A. M., Hasse, D., Valegard, K. & Andersson, I. Crystal structures of beta-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. J. Exp. Bot. 68, 3857–3867 (2017).
Chowdhury, C. et al. Selective molecular transport through the protein shell of a bacterial microcompartment organelle. Proc. Natl Acad. Sci. USA 112, 2990–2995 (2015).
Slininger Lee, M. F., Jakobson, C. M. & Tullman-Ercek, D. Evidence for improved encapsulated pathway behavior in a bacterial microcompartment through shell protein engineering. ACS Synthet. Biol. 6, 1880–1891 (2017).
Tsai, Y. et al. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLOS Biol. 5, e144 (2007).
Tanaka, S., Sawaya, M. R. & Yeates, T. O. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327, 81–84 (2010).
Iancu, C. V. et al. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372, 764–773 (2007).
Tanaka, S. et al. Atomic-level models of the bacterial carboxysome shell. Science 319, 1083–1086 (2008).
Sutter, M., Wilson, S. C., Deutsch, S. & Kerfeld, C. A. Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth. Res. 118, 9–16 (2013).
Cai, F. et al. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLOS ONE 4, e7521 (2009). This study provides evidence that the pentameric BMC-P proteins are essential for formation of an intact carboxysome shell.
Lassila, J. K., Bernstein, S. L., Kinney, J. N., Axen, S. D. & Kerfeld, C. A. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J. Mol. Biol. 426, 2217–2228 (2014).
Liberton, M., Austin, J. R. 2nd, Berg, R. H. & Pakrasi, H. B. Unique thylakoid membrane architecture of a unicellular N2-fixing cyanobacterium revealed by electron tomography. Plant Physiol. 155, 1656–1666 (2011).
Sagermann, M., Ohtaki, A. & Nikolakakis, K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl Acad. Sci. USA 106, 8883–8887 (2009). This study describes the structure of a single-layered BMC-T protein with three smaller pores around the central axis of symmetry.
Pang, A., Liang, M., Prentice, M. B. & Pickersgill, R. W. Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr. D Biol. Crystallogr. 68, 1642–1652 (2012).
Thompson, M. C., Cascio, D., Leibly, D. J. & Yeates, T. O. An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Sci. 24, 956–975 (2015).
Samborska, B. & Kimber, M. S. A. Dodecameric CcmK2 structure suggests beta-carboxysomal shell facets have a double-layered organization. Structure 20, 1353–1362 (2012).
Faulkner, M. et al. Direct characterization of the native structure and mechanics of cyanobacterial carboxysomes. Nanoscale 9, 10662–10673 (2017). This study describes the purification and structural characterization of β-carboxysomes.
Kinney, J. N., Salmeen, A., Cai, F. & Kerfeld, C. A. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J. Biol. Chem. 287, 17729–17736 (2012). This study identifies a peptide extension for targeting in β-carboxysomes and predicts counterparts in diverse BMCs.
Fan, C. et al. Short N-terminal sequences package proteins into bacterial microcompartments. Proc. Natl Acad. Sci. USA 107, 7509–7514 (2010).
Aussignargues, C., Paasch, B. C., Gonzalez-Esquer, R., Erbilgin, O. & Kerfeld, C. A. Bacterial microcompartment assembly: the key role of encapsulation peptides. Commun. Integr. Biol. 8, e1039755 (2015).
Erbilgin, O., Sutter, M. & Kerfeld, C. A. The structural basis of coenzyme a recycling in a bacterial organelle. PLOS Biol. 14, e1002399 (2016).
Cameron, J. C., Wilson, S. C., Bernstein, S. L. & Kerfeld, C. A. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 155, 1131–1140 (2013). This study describes the assembly pathway of β-carboxysomes using knockout mutants and fluorescently labelled reporters.
Ludwig, M., Sultemeyer, D. & Price, G. D. Isolation of ccmKLMN genes from the marine cyanobacterium, Synechococcus sp PCC7002 (Cyanobacteria), and evidence that CcmM is essential for carboxysome assembly. J. Phycol. 36, 1109–1118 (2000).
Emlyn-Jones, D., Woodger, F. J., Andrews, T. J., Price, G. D. & Whitney, S. M. A. Synechococcus PCC7942 ΔccmM (Cyanophyceae) mutant pseudoreverts to air growth without regaining carboxysomes. J. Phycol. 42, 769–777 (2006).
Woodger, F. J., Badger, M. R. & Price, G. D. Sensing of inorganic carbon limitation in Synechococcus PCC7942 is correlated with the size of the internal inorganic carbon pool and involves oxygen. Plant Physiol. 139, 1959–1969 (2005).
Berry, S., Fischer, J. H., Kruip, J., Hauser, M. & Wildner, G. F. Monitoring cytosolic pH of carboxysome-deficient cells of Synechocystis sp. PCC 6803 using fluorescence analysis. Plant Biol. 7, 342–347 (2005).
Pena, K. L., Castel, S. E., de Araujo, C., Espie, G. S. & Kimber, M. S. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc. Natl Acad. Sci. USA 107, 2455–2460 (2010).
Price, G. D., Howitt, S. M., Harrison, K. & Badger, M. R. Analysis of a genomic DNA region from the cyanobacterium Synechococcus sp. strain PCC7942 involved in carboxysome assembly and function. J. Bacteriol. 175, 2871–2879 (1993).
Long, B. M., Badger, M. R., Whitney, S. M. & Price, G. D. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J. Biol. Chem. 282, 29323–29335 (2007).
Long, B. M., Tucker, L., Badger, M. R. & Price, G. D. Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol. 153, 285–293 (2010).
Cot, S. S., So, A. K. & Espie, G. S. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J. Bacteriol. 190, 936–945 (2008).
McGurn, L. D. et al. The structure, kinetics and interactions of the beta-carboxysomal beta-carbonic anhydrase, CcaA. Biochem. J. 473, 4559–4572 (2016).
Zarzycki, J., Axen, S. D., Kinney, J. N. & Kerfeld, C. A. Cyanobacterial-based approaches to improving photosynthesis in plants. J. Exp. Bot. 64, 787–798 (2013).
Choudhary, S., Quin, M. B., Sanders, M. A., Johnson, E. T. & Schmidt-Dannert, C. Engineered protein nano-compartments for targeted enzyme localization. PLOS ONE 7, e33342 (2012).
Fan, C. & Bobik, T. A. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J. Bacteriol. 193, 5623–5628 (2011). This study describes encapsulation peptides in the Pdu system and shows targeting of foreign proteins.
Lawrence, A. D. et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth. Biol. 3, 454–465 (2014).
Chen, A. H., Robinson-Mosher, A., Savage, D. F., Silver, P. A. & Polka, J. K. The bacterial carbon-fixing organelle is formed by shell envelopment of preassembled cargo. PLOS ONE 8, e76127 (2013).
Cai, F., Bernstein, S. L., Wilson, S. C. & Kerfeld, C. A. Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol. 170, 1868–1877 (2016).
Iancu, C. V. et al. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396, 105–117 (2010).
Cai, F. et al. Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life 5, 1141–1171 (2015). This study provides a characterization of the protein responsible for organization of the interior of α-carboxysomes.
Chaijarasphong, T. et al. Programmed Ribosomal Frameshifting Mediates Expression of the α-Carboxysome. J. Mol. Biol. 428, 153–164 (2016).
Perlmutter, J. D., Mohajerani, F. & Hagan, M. F. Many-molecule encapsulation by an icosahedral shell. eLife 5, e14078 (2016).
Fan, C., Cheng, S., Sinha, S. & Bobik, T. A. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc. Natl Acad. Sci. USA 109, 14995–15000 (2012).
Tobimatsu, T., Kawata, M. & Toraya, T. The N-terminal regions of beta and gamma subunits lower the solubility of adenosylcobalamin-dependent diol dehydratase. Biosci. Biotechnol. Biochem. 69, 455–462 (2005).
Shibata, N. et al. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J. Biol. Chem. 285, 26484–26493 (2010).
Akita, K. et al. Purification and some properties of wild-type and N-terminal-truncated ethanolamine ammonia-lyase of Escherichia coli. J. Biochem. 147, 83–93 (2010).
Zarzycki, J., Sutter, M., Cortina, N. S., Erb, T. J. & Kerfeld, C. A. In vitro characterization and concerted function of three core enzymes of a glycyl radical enzyme — associated bacterial microcompartment. Sci. Rep. 7, 42757 (2017).
Havemann, G. D., Sampson, E. M. & Bobik, T. A. PduA is a shell protein of polyhedral organelles involved in coenzyme B-12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar typhimurium LT2. J. Bacteriol. 184, 1253–1261 (2002).
Cheng, S., Sinha, S., Fan, C., Liu, Y. & Bobik, T. A. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1,2-propanediol degradation by Salmonella. J. Bacteriol. 193, 1385–1392 (2011).
Lehman, B. P., Chowdhury, C. & Bobik, T. A. The N-terminus of the PduB protein binds the protein shell of the Pdu microcompartment to its enzymatic core. J. Bacteriol. 199, e00785-16 (2017).
Cheng, S., Fan, C., Sinha, S. & Bobik, T. A. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLOS ONE 7, e47144 (2012). This study identifies the protein responsible for NADH cofactor recycling in the PDU BMC.
Chen, P., Andersson, D. I. & Roth, J. R. The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176, 5474–5482 (1994).
Stojiljkovic, I., Baumler, A. J. & Heffron, F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J. Bacteriol. 177, 1357–1366 (1995).
Roberts, E. W., Cai, F., Kerfeld, C. A., Cannon, G. C. & Heinhorst, S. Isolation and characterization of the Prochlorococcus carboxysome reveal the presence of the novel shell protein CsoS1D. J. Bacteriol. 194, 787–795 (2012).
Sutter, M. et al. Structural characterization of a newly identified component of alpha-carboxysomes: the AAA+ domain protein CsoCbbQ. Sci. Rep. 5, 16243 (2015).
Liu, Y., Jorda, J., Yeates, T. O. & Bobik, T. A. The PduL phosphotransacylase is used to recycle coenzyme A within the Pdu microcompartment. J. Bacteriol. 197, 2392–2399 (2015).
Huseby, D. L. & Roth, J. R. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J. Bacteriol. 195, 2864–2879 (2013).
Craciun, S. & Balskus, E. P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl Acad. Sci. USA 109, 21307–21312 (2012).
Craciun, S., Marks, J. A. & Balskus, E. P. Characterization of choline trimethylamine-lyase expands the chemistry of glycyl radical enzymes. ACS Chem. Biol. 9, 1408–1413 (2014).
Kalnins, G. et al. Structure and function of CutC choline lyase from human microbiota bacterium Klebsiella pneumoniae. J. Biol. Chem. 290, 21732–21740 (2015).
LaMattina, J. W. et al. 1,2-Propanediol dehydration in Roseburia inulinivorans: structural basis for substrate and enantiomer selectivity. J. Biol. Chem. 291, 15515–15526 (2016).
Urano, N. et al. Genetic analysis around aminoalcohol dehydrogenase gene of Rhodococcus erythropolis MAK154: a putative GntR transcription factor in transcriptional regulation. Appl. Microbiol. Biotechnol. 89, 739–746 (2011).
Kataoka, M. et al. A novel NADP+-dependent L-1-amino-2-propanol dehydrogenase from Rhodococcus erythropolis MAK154: a promising enzyme for the production of double chiral aminoalcohols. Lett. Appl. Microbiol. 43, 430–435 (2006).
Land, M. et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 15, 141–161 (2015).
Nobu, M. K. et al. Phylogeny and physiology of candidate phylum 'Atribacteria' (OP9/JS1) inferred from cultivation-independent genomics. ISME J. 10, 273–286 (2016).
Biddle, A. S. et al. The complete genome sequence of Clostridium indolis DSM 755(T.). Stand. Genom. Sci. 9, 1089–1104 (2014).
Lawhon, S. D. et al. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48, 1633–1645 (2003).
Andersson, R. E. Biogenic-amines in lactic acid-fermented vegetables. Lebensmittel Wissenschaft Technol. 21, 68–69 (1988).
Collier, P. D., Cromie, D. D. O. & Davies, A. P. Mechanism of formation of chloropropanols present in protein hydrolysates. J. Am. Oil Chemists Soc. 68, 785–790 (1991).
Harvey, P. C. et al. Salmonella enterica serovar typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun. 79, 4105–4121 (2011).
Klumpp, J. & Fuchs, T. M. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153, 1207–1220 (2007).
Joseph, B. et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188, 556–568 (2006).
Maadani, A., Fox, K. A., Mylonakis, E. & Garsin, D. A. Enterococcus faecalis mutations affecting virulence in the Caenorhabditis elegans model host. Infect. Immun. 75, 2634–2637 (2007).
Bertin, Y. et al. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13, 365–377 (2011).
Srikumar, S. & Fuchs, T. M. Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol. 77, 281–290 (2011).
Thiennimitr, P. et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17480–17485 (2011).
Kendall, M. M., Gruber, C. C., Parker, C. T. & Sperandio, V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3, e00050-12 (2012).
Pitts, A. C., Tuck, L. R., Faulds-Pain, A., Lewis, R. J. & Marles-Wright, J. Structural insight into the Clostridium difficile ethanolamine utilisation microcompartment. PLOS ONE 7, e48360 (2012).
Martinez-del Campo, A. et al. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. mBio 6, e00042-15 (2015).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Lang, D. H. et al. Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: selective catalysis by FMO3. Biochem. Pharmacol. 56, 1005–1012 (1998).
Koeth, R. A. et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013).
Tang, W. H. et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 116, 448–455 (2015).
Bae, S. et al. Plasma choline metabolites and colorectal cancer risk in the Women's Health Initiative Observational Study. Cancer Res. 74, 7442–7452 (2014).
Gao, X. et al. Dietary trimethylamine N-oxide exacerbates impaired glucose tolerance in mice fed a high fat diet. J. Biosci. Bioeng. 118, 476–481 (2014).
Bhardwaj, T. & Somvanshi, P. Pan-genome analysis of Clostridium botulinum reveals unique targets for drug development. Gene 623, 48–62 (2017).
Abdul-Rahman, F., Petit, E. & Blanchard, J. L. The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J. Phylogenet. Evol. Biol. 1, 118 (2013).
Lawrence, J. G. & Roth, J. R. Evolution of coenzyme B12 synthesis among enteric bacteria: Evidence for loss and reacquisition of a multigene complex. Genetics 142, 11–24 (1996).
Kerfeld, C. A. A bioarchitectonic approach to the modular engineering of metabolism. Philos. Trans R. Soc. Lond. B Biol Sci. 372, 20160387 (2017).
Kerfeld, C. A. Plug-and-play for improving primary productivity. Am. J. Bot. 102, 1949–1950 (2015).
Parsons, J. B. et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J. Biol. Chem. 283, 14366–14375 (2008).
Bonacci, W. et al. Modularity of a carbon-fixing protein organelle. Proc. Natl Acad. Sci. USA 109, 478–483 (2012).
Baumgart, M., Huber, I., Abdollahzadeh, I., Gensch, T. & Frunzke, J. Heterologous expression of the Halothiobacillus neapolitanus carboxysomal gene cluster in Corynebacterium glutamicum. J. Biotechnol. 258, 126–135 (2017).
Lin, M. T. et al. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 79, 1–12 (2014). This study presents the first step of integrating carboxysomes into plants demonstrated by expression of β-carboxysome genes in Nicotiana chloroplasts.
Lin, M. T., Occhialini, A., Andralojc, P. J., Parry, M. A. & Hanson, M. R. A faster Rubisco with potential to increase photosynthesis in crops. Nature 513, 547–550 (2014).
Price, G. D. et al. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. J. Exp. Bot. 64, 753–768 (2013).
McGrath, J. M. & Long, S. P. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 164, 2247–2261 (2014).
Gonzalez-Esquer, C. R., Shubitowski, T. B. & Kerfeld, C. A. Streamlined construction of the cyanobacterial CO2-fixing organelle via protein domain fusions for use in plant synthetic biology. Plant Cell 27, 2637–2644 (2015). This study describes the redesign of the enzymatic core of β-carboxysomes.
Sargent, F. et al. A synthetic system for expression of components of a bacterial microcompartment. Microbiology 159, 2427–2436 (2013).
Held, M. et al. Engineering formation of multiple recombinant Eut protein nanocompartments in E. coli. Sci. Rep. 6, 24359 (2016).
Quin, M. B., Perdue, S. A., Hsu, S. Y. & Schmidt-Dannert, C. Encapsulation of multiple cargo proteins within recombinant Eut nanocompartments. Appl. Microbiol. Biotechnol. 100, 9187–9200 (2016).
Wagner, H. J., Capitain, C. C., Richter, K., Nessling, M. & Mampel, J. Engineering bacterial microcompartments with heterologous enzyme cargos. Engineer. Life Sci. 17, 36–46 (2017).
Liang, M., Frank, S., Lunsdorf, H., Warren, M. J. & Prentice, M. B. Bacterial microcompartment-directed polyphosphate kinase promotes stable polyphosphate accumulation in E. coli. Biotechnol. J. 12, 1600415 (2017).
Lee, M. J., Brown, I. R., Juodeikis, R., Frank, S. & Warren, M. J. Employing bacterial microcompartment technology to engineer a shell-free enzyme-aggregate for enhanced 1,2-propanediol production in Escherichia coli. Metab. Eng. 36, 48–56 (2016).
Jakobson, C. M., Slininger Lee, M. F. & Tullman-Ercek, D. De novo design of signal sequences to localize cargo to the 1,2-propanediol utilization microcompartment. Protein Sci. 26, 1086–1092 (2017).
Parsons, J. B. et al. Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLOS ONE 5, e14009 (2010).
Jakobson, C. M. et al. Tuning the catalytic activity of subcellular nanoreactors. J. Mol. Biol. 428, 2989–2996 (2016).
Kim, E. Y. & Tullman-Ercek, D. A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments. Biotechnol. J. 9, 348–354 (2014).
Jakobson, C. M., Kim, E. Y., Slininger, M. F., Chien, A. & Tullman-Ercek, D. Localization of proteins to the 1,2-propanediol utilization microcompartment by non-native signal sequences is mediated by a common hydrophobic motif. J. Biol. Chem. 290, 24519–24533 (2015).
Dueber, J. E. et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 27, 753–759 (2009).
Luo, L. H. et al. Identification and characterization of the propanediol utilization protein PduP of Lactobacillus reuteri for 3-hydroxypropionic acid production from glycerol. Appl. Microbiol. Biotechnol. 89, 697–703 (2011).
Toth-Petroczy, A. & Tawfik, D. S. The robustness and innovability of protein folds. Curr. Opin. Struct. Biol. 26, 131–138 (2014).
Bloom, J. D. & Arnold, F. H. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl Acad. Sci. USA 106 (Suppl. 1), 9995–10000 (2009).
Pandya, C., Farelli, J. D., Dunaway-Mariano, D. & Allen, K. N. Enzyme promiscuity: engine of evolutionary innovation. J. Biol. Chem. 289, 30229–30236 (2014).
Kinney, J. N., Axen, S. D. & Kerfeld, C. A. Comparative analysis of carboxysome shell proteins. Photosynth. Res. 109, 21–32 (2011).
Cai, F., Sutter, M., Bernstein, S. L., Kinney, J. N. & Kerfeld, C. A. Engineering bacterial microcompartment shells: chimeric shell proteins and chimeric carboxysome shells. ACS Synthet. Biol. 4, 444–453 (2015). This study demonstrates that shell proteins can be exchanged between different BMC types.
Thompson, M. C. et al. Identification of a unique Fe-S cluster binding site in a glycyl-radical type microcompartment shell protein. J. Mol. Biol. 426, 3287–3304 (2014).
Pang, A., Warren, M. J. & Pickersgill, R. W. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe-4S cluster-binding site. Acta Crystallogr. D Biol. Crystallogr. 67, 91–96 (2011).
Aussignargues, C. et al. Structure and function of a bacterial microcompartment shell protein engineered to bind a [4Fe-4S] cluster. J. Am. Chem. Soc. 138, 5262–5270 (2016). This study presents the first integration of a function, electron transfer, into a BMC shell protein.
Sturms, R., Streauslin, N. A., Cheng, S. & Bobik, T. A. In Salmonella enterica, ethanolamine utilization is repressed by 1,2-propanediol to prevent detrimental mixing of components of two different bacterial microcompartments. J. Bacteriol. 197, 2412–2421 (2015).
Heldt, D. et al. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem. J. 423, 199–207 (2009). This is the first report of an ethanol utilization microcompartment.
Pang, A., Frank, S., Brown, I., Warren, M. J. & Pickersgill, R. W. Structural insights into higher order assembly and function of the bacterial microcompartment protein PduA. J. Biol. Chem. 289, 22377–22384 (2014).
Noel, C. R., Cai, F. & Kerfeld, C. A. Purification and characterization of protein nanotubes assembled from a single bacterial microcompartment shell subunit. Adv. Mater. Interfaces 3, 1500295 (2016).
Young, E. J. et al. Engineering the bacterial microcompartment domain for molecular scaffolding applications. Front. Microbiol. 8, 1441 (2017).
Mahalik, J. P., Brown, K. A., Cheng, X. & Fuentes-Cabrera, M. Theoretical study of the initial stages of self-assembly of a carboxysome's facet. ACS Nano 10, 5751–5758 (2016).
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012).
Jorda, J., Liu, Y., Bobik, T. A. & Yeates, T. O. Exploring bacterial organelle interactomes: a model of the protein-protein interaction network in the Pdu microcompartment. PLOS Comput. Biol. 11, e1004067 (2015).
Wang, P., Lombi, E., Zhao, F. J. & Kopittke, P. M. Nanotechnology: a new opportunity in plant sciences. Trends Plant Sci. 21, 699–712 (2016).
Tsai, S. J. & Yeates, T. O. Bacterial microcompartments: insights into the structure, mechanism, and engineering applications. Prog. Mol. Biol. Transl Sci. 103, 1–20 (2011).
Shively, J. M., Ball, F., Brown, D. H. & Saunders, R. E. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182, 584–586 (1973).
Menon, B. B., Dou, Z., Heinhorst, S., Shively, J. M. & Cannon, G. C. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLOS ONE 3, e3570 (2008).
Romano, K. A., Vivas, E. I., Amador-Noguez, D. & Rey, F. E. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6, e02481-14 (2015).
Acknowledgements
This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI114975-01 with infrastructure support from the U.S. Department of Energy, Basic Energy Sciences, Contract DE-FG02-91ER20021.
Author information
Authors and Affiliations
Contributions
C.A.K., C.A., J.Z., F.C. and M.S. contributed to researching data for the article. C.A.K., C.A., J.Z., F.C. and M.S. substantially contributed to the discussion of content. C.A.K. and M.S. wrote the article and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Carbonic anhydrase
-
An enzyme that catalyses the conversion of bicarbonate to carbon dioxide (and vice versa); the several subclasses of carbonic anhydrases have distinct structural folds.
- Rubisco
-
An enzyme that fixes carbon dioxide by reacting it with ribulose-1,5-bisphosphate to create two molecules of phosphoglycerate. Form 1 Rubisco is composed of eight small and eight large subunits.
- Calvin–Benson–Bassham cycle
-
A pathway for producing phosphoglyceraldehyde from CO2.
- Superloci
-
Regions on the chromosome that contain one or more operons encoding genes for a bacterial microcompartment and ancillary proteins that support the function of the organelle.
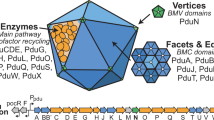
- BMC-H
-
A type of shell protein containing a single Pfam00936 domain that forms cyclic homohexamers.
- BMC-T
-
A shell protein that contains two Pfam00936 domains that form cyclic homodimers or pseudohexamers.
- BMC-P
-
A type of protein containing the Pfam03319 domain that forms homopentamers and functions as the pentagonal vertices of the BMC shell.
- Pfam00936 domain
-
An ∼90 amino acid sequence that folds into an α-β structure that oligomerizes into a hexamer.
- Facets
-
Surfaces connecting the icosahedral vertices.
- α-Carboxysomes
-
One of two types of carboxysome; α-carboxysomes encapsulate form 1A Rubisco and are found primarily in marine cyanobacteria and chemoautotrophs.
- β-Carboxysomes
-
This type of carboxysome is found in ecophysiologically diverse cyanobacteria; it encapsulates form 1B Rubisco, the form found in higher plants.
- γ-Carbonic anhydrase
-
A subclass of carbonic anhydrases with a characteristic structure of three chains that each form a left-handed β-helix and a metal ion active site.
- Encapsulation peptide
-
One or more short (∼17 amino acid) amphipathic helices that target cargo proteins to the interior of bacterial microcompartments; they are typically located at the N-terminus or C-terminus of a protein and are connected by an unstructured linker.
- Glycyl radical enzymes
-
(GREs). A class of enzymes that uses radicals of glycine and cysteine for catalysis; they are highly oxygen sensitive and require an activating enzyme containing an Fe–S cluster to generate the glycyl radical.
- Signature enzyme
-
An enzyme of a metabolosome that is specific to the initial substrate of the bacterial microcompartment.
- Maximum-likelihood tree
-
A phylogenetic tree constructed using a computationally intense method that searches for the tree that has the highest probability of producing the observed data.
- Pathogenicity islands
-
Segments of chromosomes that encode virulence factors and are found in pathogenic microorganisms but absent in closely related, non-pathogenic strains.
- Fe–S clusters
-
Metal clusters composed of non-haem iron and sulfur atoms; in proteins, they function to transfer electrons over a wide range of potentials.
- Ferredoxins
-
A family of proteins that contain Fe–S clusters to mediate electron transfer.
Rights and permissions
About this article
Cite this article
Kerfeld, C., Aussignargues, C., Zarzycki, J. et al. Bacterial microcompartments. Nat Rev Microbiol 16, 277–290 (2018). https://doi.org/10.1038/nrmicro.2018.10
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2018.10
This article is cited by
-
Structure and assembly of the α-carboxysome in the marine cyanobacterium Prochlorococcus
Nature Plants (2024)
-
Structure of intact α-carboxysome specifies role of CsoS2 in shell assembly
Nature Plants (2024)
-
Functionalization of bacterial microcompartment shell interior with cysteine containing peptides enhances the iron and cobalt loading capacity
BioMetals (2024)
-
Engineering α-carboxysomes into plant chloroplasts to support autotrophic photosynthesis
Nature Communications (2023)
-
Multiple ParA/MinD ATPases coordinate the positioning of disparate cargos in a bacterial cell
Nature Communications (2023)