Abstract
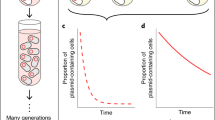
Bacteria gain antibiotic resistance genes by horizontal acquisition of mobile genetic elements (MGEs) from other lineages. Newly acquired MGEs are often poorly adapted causing intragenomic conflicts; these are resolved by either compensatory adaptation—of the chromosome or the MGE—or reciprocal coadaptation. The footprints of such intragenomic coevolution are present in bacterial genomes, suggesting an important role promoting genomic integration of horizontally acquired genes, but direct experimental evidence of the process is limited. Here we show adaptive modulation of tetracycline resistance via intragenomic coevolution between Escherichia coli and the multidrug resistant plasmid RK2. Tetracycline treatments, including monotherapy or combination therapies with ampicillin, favoured de novo chromosomal resistance mutations coupled with mutations on RK2 impairing the plasmid-encoded tetracycline efflux pump. These mutations together provided increased tetracycline resistance at reduced cost. Additionally, the chromosomal resistance mutations conferred cross-resistance to chloramphenicol. Reciprocal coadaptation was not observed under ampicillin-only or no antibiotic selection. Intragenomic coevolution can create genomes comprising multiple replicons that together provide high-level, low-cost resistance, but the resulting co-dependence may limit the spread of coadapted MGEs to other lineages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jain, R., Rivera, M. C., Moore, J. E. & Lake, J. A. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20, 1598–1602 (2003).
Frost, L. S., Leplae, R., Summers, A. O. & Toussaint, A. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732 (2005).
Norman, A., Hansen, L. H. & Sørensen, S. J. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275–2289 (2009).
Svara, F. & Rankin, D. J. The evolution of plasmid-carried antibiotic resistance. BMC Evol. Biol. 11, 130 (2011).
Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304 (2013).
Baltrus, D. A. Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495 (2013).
Diaz Ricci, J. C. & Hernández, M. E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 20, 79–108 (2000).
Harrison, E., Guymer, D., Spiers, A. J., Paterson, S. & Brockhurst, M. A. Parallel compensatory evolution stabilizes plasmids across the parasitism–mutualism continuum. Curr. Biol. 25, 2034–2039 (2015).
San Millan, A., Toll-Riera, M., Qi, Q. & MacLean, R. C. Interactions between horizontally acquired genes create a fitness cost in Pseudomonas aeruginosa. Nat. Commun. 6, 6845 (2015).
Harrison, E. & Brockhurst, M. A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267 (2012).
Porse, A., Schønning, K., Munck, C. & Sommer, M. O. A. Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873 (2016).
Loftie-Eaton, W. et al. Evolutionary paths that expand plasmid host-range: implications for spread of antibiotic resistance. Mol. Biol. Evol. 33, 885–897 (2016).
McNally, A. et al. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet. 12, e1006280 (2016).
Pansegrau, W. et al. Complete nucleotide sequence of Birmingham IncPα plasmids: compilation and comparative analysis. J. Mol. Biol. 239, 623–663 (1994).
Bottery, M. J., Wood, A. J. & Brockhurst, M. A. Selective conditions for a multidrug resistance plasmid depend on the sociality of antibiotic resistance. Antimicrob. Agents Chemother. 60, 2524–2527 (2016).
Cowan, S. W. et al. Crystal structures explain functional properties of two E. coli porins. Nature 358, 727–733 (1992).
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O. & Piddock, L. J. V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 (2015).
Cohen, S. P., McMurry, L. M., Hooper, D. C., Wolfson, J. S. & Levy, S. B. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33, 1318–1325 (1989).
Thanassi, D. G., Suh, G. S. & Nikaido, H. Role of outer membrane barrier in efflux-mediated tetracycline resistance of Escherichia coli. J. Bacteriol. 177, 998–1007 (1995).
Baba, T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006).
Lee, J., Hiibel, S. R., Reardon, K. F. & Wood, T. K. Identification of stress-related proteins in Escherichia coli using the pollutant cis-dichloroethylene. J. Appl. Microbiol. 108, 2088–2102 (2010).
Mendoza-Vargas, A. et al. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS ONE 4, e7526 (2009).
Membrillo-Hernández, J. et al. Evolution of the adhE gene product of Escherichia coli from a functional reductase to a dehydrogenase genetic and biochemical studies of the mutant proteins. J. Biol. Chem. 275, 33869–33875 (2000).
Kessler, D., Leibrecht, I. & Knappe, J. Pyruvate-formate-lyase-deactivase and acetyl-CoA reductase activities of Escherichia coli reside on a polymeric protein particle encoded by adhE. FEBS Lett. 281, 59–63 (1991).
Shasmal, M., Dey, S., Shaikh, T. R., Bhakta, S. & Sengupta, J. E. coli metabolic protein aldehyde-alcohol dehydrogenase-E binds to the ribosome: a unique moonlighting action revealed. Sci. Rep. 6, 19936 (2016).
Ma, D., Alberti, M., Lynch, C., Nikaido, H. & Hearst, J. E. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19, 101–112 (1996).
Okusu, H., Ma, D. & Nikaido, H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308 (1996).
Wang, H., Dzink-Fox, J. L., Chen, M. & Levy, S. B. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob. Agents Chemother. 45, 1515–1521 (2001).
Cooper, T. F., Rozen, D. E. & Lenski, R. E. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 1072–1077 (2003).
Hale, L., Lazos, O., Haines, A. & Thomas, C. An efficient stress-free strategy to displace stable bacterial plasmids. BioTechniques 48, 223–228 (2010).
Møller, T. S. B. et al. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. 16, 39 (2016).
Ramos, J. L. et al. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 (2005).
Allard, J. D. & Bertrand, K. P. Membrane topology of the pBR322 tetracycline resistance protein: TetA-PhoA gene fusions and implications for the mechanism of TetA membrane insertion. J. Biol. Chem. 267, 17809–17819 (1992).
Orth, P., Schnappinger, D., Hillen, W., Saenger, W. & Hinrichs, W. Structural basis of gene regulation by the tetracycline inducible Tet repressor–operator system. Nat. Struct. Mol. Biol. 7, 215–219 (2000).
Nguyen, T. N., Phan, Q. G., Duong, L. P., Bertrand, K. P. & Lenski, R. E. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol. Biol. Evol. 6, 213–225 (1989).
Stoesser, N. et al. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7, e02162-15 (2016).
Johnson, T. J. et al. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1, e00121-16 (2016).
Crozat, E., Philippe, N., Lenski, R. E., Geiselmann, J. & Schneider, D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics 169, 523–532 (2005).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6, 80–92 (2012).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Chen, K. et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods 6, 677–681 (2009).
Anderson, M. J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253 (2006).
Anderson, M. J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001).
Zapala, M. A. & Schork, N. J. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc. Natl Acad. Sci. USA 103, 19430–19435 (2006).
Acknowledgements
We thank J. P. W. Young, V. Friman, and members of the Friman and Brockhurst laboratories for discussion and comments on the manuscript. We thank C. M. Thomas for providing pCURE plasmids. This research was supported by the Wellcome Trust four-year PhD programme (WT095024MA) ‘Combating infectious disease: computational approaches in translation science’. This work was also supported by funding from the European Research Council under the European Union’s Seventh Framework Programme awarded to M.A.B. (FP7/2007-2013 ERC grant StG-2012-311490–COEVOCON).
Author information
Authors and Affiliations
Contributions
M.A.B. and A.J.W. supervised the project. M.J.B. performed the experiments and analysed the data. All authors contributed towards the design of the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information
Supplementary Figures 1–8
Supplementary Table 1
Genomic variations observed in response to experimental evolution
Rights and permissions
About this article
Cite this article
Bottery, M.J., Wood, A.J. & Brockhurst, M.A. Adaptive modulation of antibiotic resistance through intragenomic coevolution. Nat Ecol Evol 1, 1364–1369 (2017). https://doi.org/10.1038/s41559-017-0242-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41559-017-0242-3
This article is cited by
-
Mobilizable plasmids drive the spread of antimicrobial resistance genes and virulence genes in Klebsiella pneumoniae
Genome Medicine (2023)
-
Unraveling topoisomerase IA gate dynamics in presence of PPEF and its preclinical evaluation against multidrug-resistant pathogens
Communications Biology (2023)
-
Serious Risk of Tigecycline Resistance in Escherichia coli Isolated from Swine Manure
Microbial Ecology (2023)
-
Plasmid evolution in the clinic
Nature Ecology & Evolution (2022)
-
Colonization of gut microbiota by plasmid-carrying bacteria is facilitated by evolutionary adaptation to antibiotic treatment
The ISME Journal (2022)