Abstract
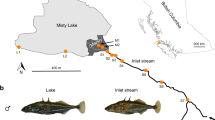
Parallel evolution of similar traits by independent populations in similar environments is considered strong evidence for adaptation by natural selection. Often, however, replicate populations in similar environments do not all evolve in the same way, thus deviating from any single, predominant outcome of evolution. This variation might arise from non-adaptive, population-specific effects of genetic drift, gene flow or limited genetic variation. Alternatively, these deviations from parallel evolution might also reflect predictable adaptation to cryptic environmental heterogeneity within discrete habitat categories. Here, we show that deviations from parallel evolution are the consequence of environmental variation within habitats combined with variation in gene flow. Threespine stickleback (Gasterosteus aculeatus) in adjoining lake and stream habitats (a lake–stream ‘pair’) diverge phenotypically, yet the direction and magnitude of this divergence is not always fully parallel among 16 replicate pairs. We found that the multivariate direction of lake–stream morphological divergence was less parallel between pairs whose environmental differences were less parallel. Thus, environmental heterogeneity among lake–stream pairs contributes to deviations from parallel evolution. Additionally, likely genomic targets of selection were more parallel between environmentally more similar pairs. In contrast, variation in the magnitude of lake–stream divergence (independent of direction) was better explained by differences in lake–stream gene flow; pairs with greater lake–stream gene flow were less morphologically diverged. Thus, both adaptive and non-adaptive processes work concurrently to generate a continuum of parallel evolution across lake–stream stickleback population pairs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Colosimo, P. F. et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005).
Losos, J. B., Jackman, T. R., Larson, A., de Queiroz, K. & Rodriguez Schettino, L. Contingency and determinism in replicated adaptive radiations of island lizards. Science 279, 2115–2118 (1998).
Langerhans, R. B. & DeWitt, T. J. Shared and unique features of evolutionary diversification. Am. Nat. 164, 335–349 (2004).
Nosil, P., Crespi, B. J. & Sandoval, C. P. Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443 (2002).
Roda, F. et al. Genomic evidence for the parallel evolution of coastal forms in the Senecio lautus complex. Mol. Ecol. 22, 2941–2952 (2013).
Schluter, D., Clifford, E. A., Nemethy, M. & McKinnon, J. S. Parallel evolution and inheritance of quantitative traits. Am. Nat. 163, 809–822 (2004).
Huss, M., Howeth, J. G., Osterman, J. I. & Post, D. M. Intraspecific phenotypic variation among alewife populations drives parallel phenotypic shifts in bluegill. Proc. Biol. Sci. 281, 20140275 (2014).
McPhail, J. D. Ecology and evolution of sympatric sticklebacks (Gasterosteus): origin of the species pairs. Can. J. Zool. 71, 515–523 (1993).
Johnson, L. S. & Taylor, E. B. The distribution of divergent mitochondrial DNA lineages of threespine stickleback (Gasterosteus aculeatus) in the northeastern Pacific Basin: post-glacial dispersal and lake accessibility. J. Biogeogr. 31, 1073–1083 (2004).
Caldera, E. J. & Bolnick, D. I. Effects of colonization history and landscape structure on genetic variation within and among threespine stickleback (Gasterosteus aculeatus) populations in a single watershed. Evol. Ecol. Res. 10, 575–598 (2008).
Deagle, B. E. et al. Population genomics of parallel phenotypic evolution in stickleback across stream–lake ecological transitions. Proc. Biol. Sci. 279, 1277–1286 (2012).
Deagle, B. E., Jones, F. C., Absher, D. M., Kingsley, D. M. & Reimchen, T. E. Phylogeography and adaptation genetics of stickleback from the Haida Gwaii archipelago revealed using genome-wide single nucleotide polymorphism genotyping. Mol. Ecol. 22, 1917–1932 (2013).
Kaeuffer, R., Peichel, C. L., Bolnick, D. I. & Hendry, A. P. Parallel and nonparallel aspects of ecological, phenotypic, and genetic divergence across replicate population pairs of lake and stream stickleback. Evolution 66, 402–218 (2012).
Berner, D., Adams, D. C., Grandchamp, A.-C. & Hendry, A. P. Natural selection drives patterns of lake–stream divergence in stickleback foraging morphology. J. Evol. Biol. 21, 1653–1665 (2008).
Mahler, D. L., Ingram, T., Revell, L. J. & Losos, J. B. Exceptional convergence on the macroevolutionary landscape in island lizard radiations. Science 341, 292–295 (2013).
Marchinko, K. B., Matthews, B., Arnegard, M. E., Rogers, S. M. & Schluter, D. Maintenance of a genetic polymorphism with disruptive natural selection in stickleback. Curr. Biol. 24, 1289–1292 (2014).
Kitano, J. et al. Reverse evolution of armor plates in the threespine stickleback. Curr. Biol. 18, 769–774 (2008).
Bańbura, J. Lateral plate morph differentiation of freshwater and marine populations of the three-spined stickleback, Gasterosteus aculeatus, in Poland. J. Fish Biol. 44, 773–783 (1994).
Leinonen, T., McCairns, R. J. S., Herczeg, G. & Merilä, J. Multiple evolutionary pathways to decreased lateral plate coverage in freshwater threespine sticklebacks. Evolution 66, 3866–3875 (2012).
Gould, S. J. Wonderful Life: The Burgess Shale and the Nature of History (W. W. Norton & Co., 1989).
Nosil, P. & Crespi, B. J. Does gene flow constrain adaptive divergence or vice versa? A test using ecomorphology and sexual isolation in Timema cristinae walking-sticks. Evolution 58, 102–112 (2004).
Kolbe, J. J., Leal, M., Schoener, T. W., Spiller, D. A. & Losos, J. B. Founder effects persist despite adaptive differentiation: a field experiment with lizards. Science 335, 1086–1089 (2012).
Lucek, K., Lemoine, M. & Seehausen, O. Contemporary ecotypic divergence during a recent range expansion was facilitated by adaptive introgression. J. Evol. Biol. 27, 2233–2248 (2014).
Theis, A., Ronco, F., Indermaur, A., Salzburger, W. & Egger, B. Adaptive divergence between lake and stream populations of an East African cichlid fish. Mol. Ecol. 23, 5304–5322 (2014).
Berner, D., Grandchamp, A.-C. & Hendry, A. P. Variable progress toward ecological speciation in parapatry: stickleback across eight lake-stream transitions. Evolution 63, 1740–1753 (2009).
Hanifin, C. T., Brodie, E. D. & Brodie, E. D. Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 6, e60 (2008).
Heinen, J. L. et al. Environmental drivers of demographics, habitat use, and behavior during a post-Pleistocene radiation of Bahamas mosquitofish (Gambusia hubbsi). Evol. Ecol. 27, 971–991 (2013).
Ravinet, M., Prodöhl, P. A. & Harrod, C. Parallel and nonparallel ecological, morphological and genetic divergence in lake-stream stickleback from a single catchment. J. Evol. Biol. 26, 186–204 (2013).
Adams, D. C. & Collyer, M. L. A general framework for the analysis of phenotypic trajectories in evolutionary studies. Evolution 63, 1143–1154 (2009).
Hendry, A. P. & Taylor, E. B. How much of the variation in adaptive divergence can be explained by gene flow? An evaluation using lake-stream stickleback pairs. Evolution 58, 2319–2331 (2004).
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37135 (2012).
Oke, K. B. et al. Does plasticity enhance or dampen phenotypic parallelism? A test with three lake-stream stickleback pairs. J. Evol. Biol. 29, 126–143 (2015).
Zelditch, M. L ., Swiderski, D. L & Sheets, H. D. Geometric Morphometrics for Biologists: A Primer (Elsevier, 2012).
Adams, D. C. & Otárola-Castillo, E. Geomorph: an rpackage for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 4, 393–399 (2013).
Albert, A. Y. K. et al. The genetics of adaptive shape shift in stickleback: pleiotropy and effect size. Evolution 62, 76–85 (2008).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Snowberg, L. K., Hendrix, K. M. & Bolnick, D. I. Covarying variances: more morphologically variable populations also exhibit more diet variation. Oecologia 178, 89–101 (2015).
Rohland, N. & Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946 (2012).
Catchen, J., Hohenlohe, P. A., Bassham, S., Amores, A. & Cresko, W. A. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22, 3124–3140 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Lleonart, J., Salat, J. & Torres, G. J. Removing allometric effects of body size in morphological analysis. J. Theor. Biol. 205, 85–93 (2000).
Acknowledgements
We thank B. Anholt, O. Banjoko, M. Dubin, L. Duncan, S. Halbrook, T. Ingram, A. Kamath, E. Delaney, C. LeBlond, J. Losos, K. Oke, S. Pakula, R. Rangel, S. Rogers, G. Rolshausen, S. Rudman, O. Schmidt, W. Shim, C. Tanner, L. Tanter, the Schluter Lab, the Bamfield Marine Sciences Centre, the Genome Sequencing and Analysis Facility at UT Austin and the Texas Advance Computing Center at UT Austin for their help, advice and comments throughout the research and writing. The British Columbia Ministry of the Environment provided essential permits. The research was supported by National Science Foundation grants DEB-1144773 (D.I.B. and A.P.H.), DEB-1144556 (C.L.P.) and IOS-1145468 (D.I.B.) and conducted in full compliance with ethical regulations according to UT Austin’s Institutional Care and Use Committee (AUP-2012-00065 and AUP-2014-00293).
Author information
Authors and Affiliations
Contributions
A.P.H., C.L.P., R.D.H.B., D.H., B.K.L., Y.E.S. and D.I.B. planned the study. All authors executed the study. Y.E.S. oversaw field collections, conducted with D.I.B., C.L.P., A.P.H., R.D.H.B., D.H., B.K.L., T.T., A.D. and R.I. Y.E.S. oversaw morphological and environmental data collections by Y.E.S., D.I.B., C.J.T., T.T., N.A. and R.I. Y.E.S. collected genomic data with help from J.N.W. and D.I.B. Y.E.S., T.V. and D.I.B. analysed the data. M.R. estimated the population genetic parameters. All authors participated in the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Descriptions of approximate Bayesian computation analyses, phenotype–environment analyses, Supplementary Tables 1–8, and Supplementary Figures 1–6. (PDF 1758 kb)
Supplementary Dataset
Pair-by-trait, lake–stream t-tests. Each row represents the multidimensional lake–stream divergence vector for a given pair. Each column is the t-statistic from a t-test comparing lake versus adjoined stream for that trait. (CSV 9 kb)
Rights and permissions
About this article
Cite this article
Stuart, Y., Veen, T., Weber, J. et al. Contrasting effects of environment and genetics generate a continuum of parallel evolution. Nat Ecol Evol 1, 0158 (2017). https://doi.org/10.1038/s41559-017-0158
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41559-017-0158
This article is cited by
-
Armour reduction and pelvic girdle loss in a population of threespine stickleback (Gasterosteus aculeatus) from western Newfoundland, Canada
Environmental Biology of Fishes (2023)
-
Variation in morphology among populations of threespine stickleback (Gasterosteus aculeatus) from western Newfoundland, Canada
Environmental Biology of Fishes (2023)
-
Climatic similarity and genomic background shape the extent of parallel adaptation in Timema stick insects
Nature Ecology & Evolution (2022)
-
Non-parallel morphological divergence following colonization of a new host plant
Evolutionary Ecology (2022)
-
Ecology directs host–parasite coevolutionary trajectories across Daphnia–microparasite populations
Nature Ecology & Evolution (2021)