Abstract
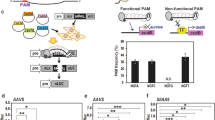
Clustered regularly interspaced short palindromic repeats (CRISPR)–Cpf1 has emerged as an effective genome editing tool in animals. Here we compare the activity of Cpf1 from Acidaminococcus sp. BV3L6 (As) and Lachnospiraceae bacterium ND2006 (Lb) in plants, using a dual RNA polymerase II promoter expression system. LbCpf1 generated biallelic mutations at nearly 100% efficiency at four independent sites in rice T0 transgenic plants. Moreover, we repurposed AsCpf1 and LbCpf1 for efficient transcriptional repression in Arabidopsis, and demonstrated a more than tenfold reduction in miR159b transcription. Our data suggest promising applications of CRISPR–Cpf1 for editing plant genomes and modulating the plant transcriptome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Carroll, D. Genetics 188, 773–782 (2011).
Bogdanove, A. J. & Voytas, D. F. Science 333, 1843–1846 (2011).
Paul, J. W. III & Qi, Y. Plant Cell Rep. 35, 1417–1427 (2016).
Zetsche, B. et al. Cell 163, 759–771 (2015).
Kim, Y. et al. Nat. Biotechnol. 34, 808–810 (2016).
Hur, J. K. et al. Nat. Biotechnol. 34, 807–808 (2016).
Port, F. & Bullock, S. L. Nat. Methods 13, 852–854 (2016).
Gao, Y. & Zhao, Y. J. Integr. Plant Biol. 56, 343–349 (2014).
Kleinstiver, B. P. et al. Nat. Biotechnol. 34, 869–874 (2016).
Kim, D. et al. Nat. Biotechnol. 34, 863–868 (2016).
Lowder, L. G. et al. Plant Physiol. 169, 971–985 (2015).
Xu, R. et al. Plant Biotechnol. J. http://doi.org/bzh2 (2016).
Endo, A., Masafumi, M., Kaya, H. & Toki, S. Sci. Rep. 6, 38169 (2016).
Tang, X. et al. Mol. Plant 9, 1088–1091 (2016).
Murray, M. G. & Thompson, W. F. Nucleic Acids Res. 8, 4321–4325 (1980).
Magoc, T. & Salzberg, S. L. Bioinformatics 27, 2957–2963 (2011).
Li, H. & Durbin, R. Bioinformatics 26, 589–595 (2010).
Acknowledgements
This work was supported by grants including the National Science Foundation of China (31330017, 31271420 and 31371682) and the National Transgenic Major Project (2014ZX0801003B-002) to X.Z. and Y.Z., a Collaborative Funding Grant from North Carolina Biotechnology Center and Syngenta Biotechnology (2016-CFG-8003) and a start-up fund from University of Maryland-College Park to Y.Q., and National Science Foundation (MCB 0209818, DBI 0923827 and 105-1339209) to D.F.V.
Author information
Authors and Affiliations
Contributions
Y.Q., Y.Z. and D.F.V. designed the experiments. Y.Q., L.G.L. and A.A.M. generated all the constructs. X.T. and Y.Z. performed the transient assays in protoplasts and prepared samples for deep sequencing. T.Z., Y.Z. and X.T. analysed the deep sequencing data. X.T., X.Z., Z.Z., Y.C., Q.R. and Q.L. generated stable transgenic rice and analysed the plants. L.G.L. and E.R.K. produced Arabidopsis transcriptional repression data. Y.Q., Y.Z. and D.F.V. wrote the paper with input from other authors. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary References, Supplementary Figures 1–10, Supplementary Table 1 (Oligos and gBlocks used in this study). (PDF 3235 kb)
Supplementary Table 2
Assembled T-DNA vectors used in this study. (XLSX 11 kb)
Supplementary Table 3
Sample information for high-throughput sequencing. (XLS 14 kb)
Rights and permissions
About this article
Cite this article
Tang, X., Lowder, L., Zhang, T. et al. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nature Plants 3, 17018 (2017). https://doi.org/10.1038/nplants.2017.18
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nplants.2017.18
This article is cited by
-
Genome editing based trait improvement in crops: current perspective, challenges and opportunities
The Nucleus (2024)
-
Efficient genome editing in grapevine using CRISPR/LbCas12a system
Molecular Horticulture (2023)
-
Targeted mRNA demethylation in Arabidopsis using plant m6A editor
Plant Methods (2023)
-
Efficient plant genome engineering using a probiotic sourced CRISPR-Cas9 system
Nature Communications (2023)
-
An efficient CRISPR–Cas12a promoter editing system for crop improvement
Nature Plants (2023)